Abstract
Lappula myosotis V. Wolf 1776 is an annual or biennial plant with important medicinal value. In the present study, we report the complete chloroplast genome data of L. myosotis, which has a length of 146,668 bp, including a small single-copy (SSC) region of 17,059 bp, a large single-copy (LSC) region of 79,691 bp, and a pair of inverted repeats (IRs) of 24,959 bp. A total of 127 genes encoding tRNA and rRNA were annotated. The total CG content of the chloroplast genome was 37.7%. The maximum-likelihood (ML) phylogenetic tree strongly supported that L. myosotis is closely related to Trigonotis peduncularis. The complete chloroplast genome of L. myosotis provides useful information on the evolution and phylogenetic relationship among Boraginaceae plants.
Introduction
Lappula myosotis V. Wolf 1776 is an annual or biennial plant of the genus Lappula in the family Boraginaceae. L. myosotis has important medicinal value and can be anti-inflammatory and insecticidal (Zhang et al. Citation2005). L. myosotis grows in grassland, hillside grassland, etc. The plant species were distributed in North China, Northwest China, and Western Inner Mongolia as well as central and Eastern Europe, North America, Afghanistan, and Pakistan (Wang et al. Citation1986). To better understand the genomic structure of L. myosotis and its phylogenetic position in Boraginaceae, we sequenced the complete chloroplast genome of L. myosotis and compared it with its close relatives.
Materials
We collected samples of L. myosotis in Heilongjiang Province, China, in June 2021 (N 44°60′16″, E 129°75′69″) (). Voucher specimens were deposited in the Herbarium of Heilongjiang University of Chinese Medicine (registration number: MDJ20210607001) (Dr. Yan, [email protected]). DNA samples were stored in the Molecular Laboratory of Heilongjiang University of Chinese Medicine (Harbin, China).
Methods
DNA was extracted from fresh leaves of L. myosotis by CTAB (Hamad Citation2011). Genomic DNA was sequenced using an Illumina HiSeq (Benagen Co., Wuhan, China) with paired-end (PE) sequencing. The chloroplast genome of Lithospermum erythrorhizon (NC053783) served as our reference genome. Sequencing raw data quality was assessed using FastQC v0.11.7 software (de Sena Brandine and Smith Citation2019). GetOrganelle V1.7.5 (Daniell et al. Citation2016) software was used to splice the chloroplast genome, and GeSeq (Tillich et al. Citation2017) software was used for functional annotation. Quantities of rRNA and tRNA were checked using the tRNAscanSE tool (Chan et al. Citation2021). Finally, RAxML v8.2.12 (Stamatakis Citation2014) was used to construct a maximum-likelihood (ML) tree, and bootstrap values were based on 1000 replicates. The complete chloroplast genome sequence of L. myosotis was submitted to GenBank with accession number MZ959108.
Results
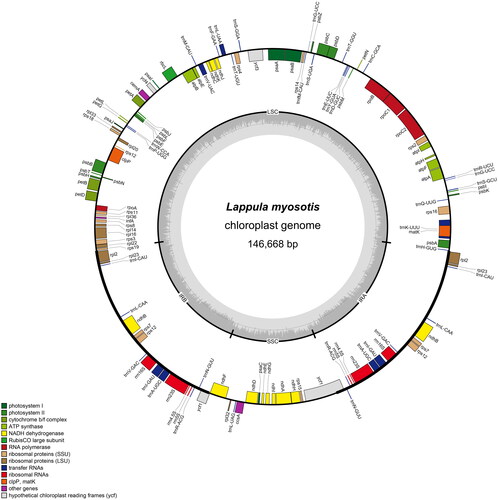
The chloroplast genome length of L. myosotis is 146,668 bp, which includes a large single-copy (LSC) region of 79,691 bp, a small single-copy (SSC) region of 17,059 bp and a pair of inverted repeats (IRs) of 24,959 bp (). The total nucleotide composition was 30.7% A, 31.5% T, 19.2% C, and 18.5% G. A total of 127 genes were annotated, including 83 protein coding genes, 36 tRNAs, and eight rRNAs.
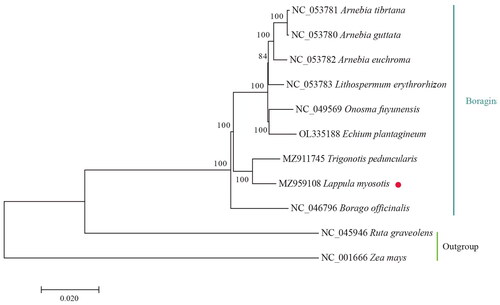
To validate the phylogenetic position of L. myosotis, eight species from Boraginaceae and two species from Rutaceae (Ruta graveolens) and Poales (Zea mays) as an outgroup were used to construct a ML phylogenetic tree with 1000 bootstrap replications (Best-fit model: GTR + G) (). The following sequences were used: Arnebia tibetana (NC_053781), Arnebia guttata (NC_053780), Arnebia euchroma (NC_053782), Lithospermum erythrorhizon (NC_053783), Onosma fuyunensis (NC_049569) (Yi et al. Citation2021), Echium plantagineum (OL335188) (Inês et al. Citation2022), Trigonotis peduncularis (MZ911745), Borago officinalis (NC_046796) (Zhen and Zhang Citation2019), and Zea mays (NC_001666) (Maier et al. Citation1995). The sequences were aligned using MAFFT (version: 7.313). Phylogenetic analysis showed that L. myosotis is most closely associated with Trigonotis peduncularis.
Discussion and conclusions
The complete chloroplast genome of L. myosotis is the first report of a member of Lappula, which fills the gap in genome-related information. Provide data support for the subsequent classification of Boraginaceae.
Author contributions
SD, SY, and TW conceived the research. ZW, WM, WR, and CL designed the experiment, data analysis, and wrote the paper. SD and WM revised the paper. All authors contributed to the article and approved the submitted version.
Ethical approval
No ethical approval/permission is required in this study. This study includes no human, animal, or endangered plant samples, and the sample was legally collected in accordance with guidelines provided by the authors’ institution and national or international regulations.
Disclosure statement
The authors have declared that no competing interests exist.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MZ959108. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA758790, SRR15666708, and SAMN21035694, respectively.
Correction Statement
This article has been corrected with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Chan PP, Lin BY, Mak AJ, Lowe TM. 2021. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 49(16):9077–9096.
- Daniell H, Sea Lin C, Yu M, Chang WJ. 2016. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17(1):1–29.
- de Sena Brandine G, Smith AD. 2019. Falco: high-speed FastQC emulation for quality control of sequencing data. F1000Res. 8:1874.
- Hamad AI. 2011. Modified CTAB method for high quality genomic DNA extraction from medicinal plants. Pak J Biol Sci. 14(21):998–999.
- Inês CL, Teresa BCM, Jorge C, Frédéric BG. 2022. The complete plastome of Echium plantagineum L. (Boraginaceae), the first chloroplast genome belonging to the Echium genus. Mitochondrial DNA B Resour. 7(6):1154–1156.
- Maier RM, Neckermann K, Igloi GL, Kössel H. 1995. Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J Mol Biol. 251(5):614–628.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wang XC, Pei YH, Li X, Zhu TR. 1986. Studies on the antibacterial constituents in the fruit of Lappula echinata Gilib. Yao Xue Xue Bao = Acta Pharm Sin. 21(3):183–186.
- Yi H, Xuemin X, Quanru L. 2021. The complete chloroplast genome of Onosma fuyunensis Y.He & Q.R. Liu and its phylogenetic analysis. Mitochondrial DNA B Resour. 6(11):3142–3143.
- Zhang S-Y, Meng L, Gao W-Y, Jia W, Duan H-Q. 2005. A new quinolone alkaloid with antibacterial activity from Lappula echinata. Chin Tradit Herb Drugs. 36(4):490–492.
- Zhen LL, Zhang SD. 2019. The complete chloroplast genome of the multipurpose and traditional herb, Ruta graveolens L. Mitochondrial DNA B Resour. 4(2):3661–3662.