Abstract
The Brambling (Fringilla montifringilla) (Linnaeus 1758) is a member of the Passeriformes family of birds and primarily feeds on grass seeds and grains. Muscle tissue was collected from birds sampled from Moar Mountain, China, and the complete mitochondrial genome was sequenced. Its mitochondrial genome consists of 13 protein-coding genes (PCGs), 2 rRNA genes (12S rRNA and 16S rRNA), 22 tRNA genes, and 1 control region (CR). The genome comprises 30.30% A, 23.32% T, 14.31% G, and 32.07% C bases. Phylogenetically, F. montifringilla is closely related to the Fringilla coelebs, Fringilla teydea teydea and Fringilla polatzeki.
Introduction
The Brambling (Fringilla montifringilla) (Linnaeus 1758), a member of the Passeriformes family of birds, a very large and diverse family commonly known as perching birds, and is also known as the Tiger-skin Brambling (Liu et al. Citation2004; Fang et al. Citation2008). Large population numbers occupy a very wide geographical range and inhabit large areas of the northern Asian continent (MacKinnon and Phillipps Citation2002), primarily feeding on grass seeds and grains (Liu et al. Citation2004). Although the population size is declining, the rate of decline is slow, and it is classified as a species of Least Concern on the Red Data List (Vikan et al. Citation2010).
Currently, F. montifringilla research focuses on energetics and thermoregulation (Liu et al. Citation2004), vocalization (Jiang et al. Citation2001; Zhao et al. Citation2003), sex ratio and age composition (Liu and Liu Citation2021; Jenni Citation2022; Kumar et al., Citation2016), migration (Fang et al. Citation2008), and evolution of defenses (Vikan et al. Citation2010). Evolutionary analysis and data on the species’ mitochondrial genome are limited. To date, more than 569 mitochondrial genomes have been sequenced and stored on Genbank as part of the BioProject initiative. In this study, we sequenced the complete mitochondria of F. montifringilla and analyzed its phylogenic relationship to other birds of the same genus. This data is of reference significance for the protection of and subsequent research on the Brambling.
Materials and methods
The mitochondrial genome of F. montifringilla was sequenced from muscle tissue collected from Maor Mountain in China (127°30′–127°34′E, 45°20′–45°25′N). After use, specimens were deposited at Northeast Forestry University’s College of Wildlife and Protected Areas, voucher number YQ202106 (Zhensheng Liu, Email: zhenshengliu @163.com) ().
Figure 1. The F. montifringilla reference image. This image was taken by Dr. Meng Dehuai using a Canon 700 D camera in Maor Mountain, China (127°30′–127°34′E, 45°20′–45°25′N).

Illumina HiSeq sequencing technology was used to perform paired-end (PE) sequencing using the whole-genome shotgun (WGS) method and next-generation sequencing (NGS). DNA was extracted from samples, followed by purification, database building, and sequencing, and the resulting readable Raw Data was uploaded to GenBank as SRR17012662. The complete genome was submitted to GenBank under accession number OL629048.1.
A maximum likelihood analysis was performed in MEGA7 (Kumar et al. Citation2016) to infer phylogenetic relationships (Rogers and Swofford Citation1998; Chen et al. Citation2022).
Results
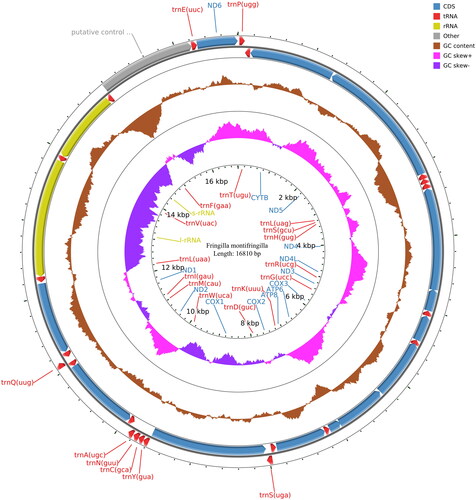
The mitochondrial genome of F. montifringilla consists of 13 protein-coding genes (PCGs), 2 rRNA genes (12S rRNA and 16S rRNA), 22 tRNA genes, and 1 control region (CR). The genome is 16,810 bp long, with a GC content of 46.38%. The genome consists of 30.30% A, 23.32% T, 14.31% G, and 32.07% C bases. The total length of the 13 protein-coding genes is 11,413 bp. Besides the initiation codon, 11 are ATG (ND1, ND2, ND4, ND4L, ND5, ND6, COX2, COX3, ATP6, ATP8, and CYTB), one is ATT (ND3), and one is GTG (COX1). There were nine stop codons with TAA (ND2, ND3, ND4, ND4L, COX2, COX3, ATP6, ATP8, and CYTB), two with AGG (COX1, ND1), one with AGA, and one with TAG (ND6, ND5). All the tRNAs comprised 1562 base pairs. The two rRNA genes comprised 1598 base pairs (12 s rRNA) and 976 base pairs (16 s rRNA), with a control region of 1233 base pairs ().
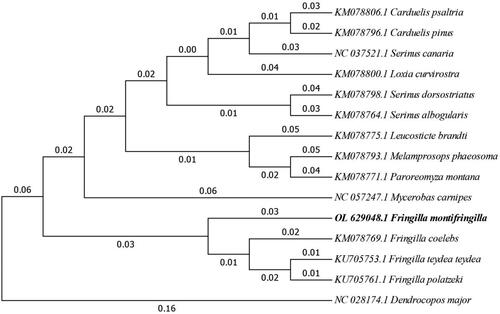
The bootstrap consensus tree inferred from 1000 replicates is used to represent the evolutionary history of the taxa analyzed (Felsenstein Citation1985). As shown in the bootstrap consensus tree (), F. montifringilla is closely related to the Fringilla coelebs, Fringilla teydea teydea and Fringilla polatzeki.
Discussion
The population size of F. montifringilla is decreasing, although slowly (Lindstrom Citation1987). This is possibly due to the breeding site tenacity of this species, where ringing recovery studies indicate that the Bramblings breed at sites that are up to 600 km apart in different years (Mikkonen Citation1983). Despite this tenacity, it is important to sequence the complete mitochondrial genome of this species for reference significance and the protection of subsequent research on the Brambling.
In recent years, research on the phylogeny and evolution of passerines has attracted much attention. For example, Bayesian analysis was used to study the taxonomic status of this subfamily, Subfamily name, of birds revealing a heterogeneous origin of the Fringilla genus (Antonio et al. Citation2007). Additionally, mitochondrial data is often used to infer phylogenetic relationships in these birds. Guo et al. (Citation2007) analyzed the mitochondrial cytochrome b gene sequence of 18 passerine birds to infer their phylogeny. The mitochondrial genome sequence of F. montifringilla obtained in this study will provide useful genetic data for further phylogenetic and evolutionary analysis.
Ethical approval
The study was approved by the institutional review board of Northeast Forestry University, Heilongjiang, China. The collection of bird muscle tissue was conducted following the guidelines provided by the Northeast Forestry University and the Heilongjiang province regulations under reference number YQ202106. Field studies complied with Heilongjiang province legislation.
Author contributions
Thank you to Professors Zhensheng Liu and Liwei Teng for their contributions to the study concept and design. We are grateful to Dr. Wei Yu, Dr. Dehuai Meng, Junda Chen, Xu Zhang, and Xuyang Zhao for the sample collection. Dr. Cui Shuang’s contribution to the conception and design of the thesis, analysis, and interpretation of the data, and the drafting of the paper is appreciated. All authors agree to be accountable for all aspects of the work.
Disclosure statement
The authors report no conflicts of interest. They also declare that they have no known competing financial interests or personal relationships that could influence the work reported in this paper.
Data availability statement
The data supporting this study’s findings are freely available on the NCBI website at https://www.ncbi.nlm.nih.gov/, reference number OL629048.1. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA782766, SRR17012662, and SAMN23394671, respectively.
Additional information
Funding
References
- Antonio A, Juan M, Valentin R, Javier G, Raquel R, Michael W, Juan IS. 2007. Bayesian phylogeny of Fringillinae birds: status of the singular African oriole finch Linurgus olivaceus and evolution and heterogeneity of the genus Carpodacus. Acta Zool Sinica. 53(5):826–834.
- Chen JD, Meng DH, Si YH, Yu MC, Teng LW, Liu ZS, Zhou XY. 2022. Characterization of the complete mitochondrial genome of the lesser spotted woodpecker (Dryobates minor) and its phylogenetic position. Mitochondrial DNA Part B. 7(8):1504–1506.
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 39(4):783–791.
- Fang KJ, Li XD, Guo YN, Li F, Yu XD. 2008. Migration of brambling (Fringilla montifringilla) in Gaofeng Forestry Area of Nenjiang District. Chin J Wildlife. 29(3):121–123.
- Guo HY, Li L, Ma YK, Bai SY. 2007. Phylogenetic relationship among 18 passerines based on mitochondrial cytochrome b gene sequences. Chin J Zool. 42(2):32–38.
- Jiang J, Li D, Li J, Yang X. 2001. Effect of lesion of nucleus robustus archistriatalis on call in bramble finch (Fringilla montifringilla). Sci China C Life Sci. 44(5):479–488.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Jenni L. 2022. Which birds participate in mass concentrations of bramblings Fringilla montifringilla? Ring recoveries, biometry, age and sex composition. J Ornithol. 163(1):1–17.
- Lindstrom A. 1987. Breeding nomadism and site tenacity in the brambling Fringilla montifringilla. Ornis Fenn. 64:50–56.
- Liu JS, Song CG, Wang XH, Chen MH, Wang Y. 2004. A comparison of metabolic theromogenesis and digestive tract morphology between bramblings and tree sparrows. Chin J Zool. 39(3):2–7.
- Liu JS, Wang DH, Wang Y, Chen MH, Song CG, Sun RY. 2004. Energetics and thermoregulation of the Carpodacus roseus, Fringilla montifringilla and Acanthis flammea. Acta Zool Sinica. 50(3):357–363.
- Liu LP, Liu PF. 2021. Male-biased sex ratio and body size of a wintering roost of brambling, Fringilla montifringilla Linnaeus, 1758. PJZ. 53(6):2479–2482.
- Mikkonen AV. 1983. Breeding site tenacity of the chaffinch Fringilla coelebs and the brambling F. montifringilla in northern Finland. Ornis Scand. 14(1):36–47.
- MacKinnon J, Phillipps K. 2002. A field guide to the birds of China. Oxford: Oxford University Press.
- Rogers JS, Swofford DL. 1998. A fast method for approximating maximum likelihoods of phylogenetic trees from nucleotide sequences. Syst Biol. 47(1):77–89.
- Vikan JR, Stokke BG, Rutila J, Huhta E, Moksnes A, Roskaft E. 2010. Evolution of defences against cuckoo (Cuculus canorus) parasitism in bramblings (Fringilla montifringilla): a comparison of four populations in Fennoscandia. Evol Ecol. 24(5):1141–1157.
- Zhao J, Jiang J, Li DF. 2003. Control pattern of nucleus dorsalis medialis of the intercollicular comrlex (DM) for vocalizatlon in bramble finch (Fringilla montifringilla). Acta Biophysica Sinica. 19(2):151–155.