Abstract
Tulipa sinkiangensis Z. M. Mao 1980 is endemic to Xinjiang Province, China. In this study, we reported the complete chloroplast genome of T. sinkiangensis. The complete chloroplast genome of T. sinkiangensis comprises 151,929 bp and was divided into four typical regions: a large single-copy region of 82,062 bp, a pair of inverse repeats of 26,361 bp each, and a small single-copy region of 17,145 bp. A total of 136 genes were identified in this chloroplast, of which 87 were protein-coding, 38 were tRNA, eight were rRNA, and three were pseudogene. The results of this study will provide valuable information for understanding evolution and identification of different species belonging to genus Tulipa.
Keywords:
Tulipa L. is a genus of perennial herbaceous plant in the Liliaceae family. The number of species in this genus ranges from about 45 (Stork Citation1984) to more than 130 (Diana Citation2013). The Pamir Alai and Tianshan Mountains are primary gene resource centers for plants belonging to genus Tulipa (Hoog Citation1973). More than 10% Tulipa species were reported as being native to China (Tang and Wang Citation1980; Han Citation2014). Germplasm resources have been investigated in China (Jiao et al. Citation2015). Such studies have concentrated on describing its morphological variation (Chen et al. Citation2001; Mei and Tan Citation2006; Jiao et al. Citation2015). In recent years, the chloroplast genome has been used to study the genetic diversity and phylogenetic relationship in Tulipa species (Zhou et al. Citation2019; Ju, Shi, et al. Citation2020; Ju, Tang, et al. Citation2020). In this study, we worked on T. sinkiangensis Z. M. Mao 1980 which is a endemic species in China. The aim is to provide the complete chloroplast genome sequence of T. sinkiangensis. This is a necessary process to comprehend its phylogenetic relationships with other Tulipa species. Samples of T. sinkiangensis used in this study were collected from the low mountain slopes around Urumqi city (87°34′E, 43°47′ N, 924 m altitude, Xinjiang Province, China). The corresponding voucher specimen (no. 15006) was deposited in the Liaoning Academy of Agricultural Sciences (https://www.laas.cn/; the contact person is Huihua Zhang, [email protected]).
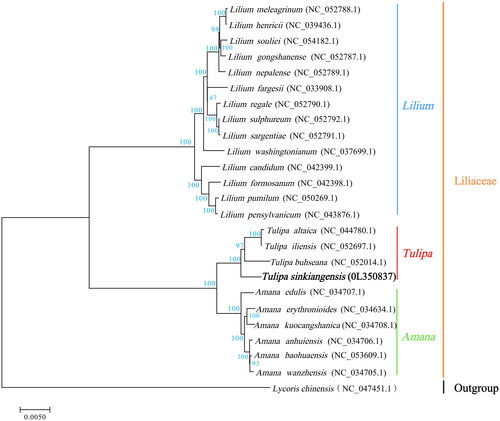
The total DNA of T. sinkiangensis was extracted from its leaves using the modified CTAB method (Doyle Citation1987; Yang et al. Citation2014). Sequencing libraries were generated using a TruSeq DNA Sample Preparation Kit (Illumina, San Diego, CA). Genome sequencing was performed using Illumina NovaSeq platform. The filtered reads were assembled by SPAdes v3.10.1 (Bankevich et al. Citation2012) and the assembled genome was annotated by an integrated plastome sequence annotator CPGAVAS2 (Shi et al. Citation2019). The chloroplast genome sequence of T. sinkiangensis was submitted to the NCBI database with the accession number OL350837. Complete chloroplast genome had a typical quadripartite structure, which was 151,929 bp in length with GC content of 36.69%. It contained a large single-copy region (LSC) of 82,062 bp, a small single-copy region (SSC) of 17,145 bp, and two inverted repeat (IR) regions of 26,361 bp. A total of 136 genes were successfully annotated, including 87 protein-codon genes, 38 tRNA genes, eight rRNA genes, and three pseudogenes. To understand the phylogenetic position of T. sinkiangensis with its close relatives, phylogenetic analysis was performed based on 24 complete chloroplast genomes of various species in Liliaceae. These genomes were aligned by MAFFT v7.427 (Katoh and Standley Citation2013). Evolution history was inferred by using maximum-likelihood method based on Tamura–Nei model in RAxML v8.2.10. Bootstrap value was calculated based on 1000 replicate analysis. Results showed that T. sinkiangensis, T. buhseana, T. altaica, and T. iliensis were clustered in a unique clade in the Tulipa genus ().
Author contributions
The article was designed and conceived by Lianwei Qu and Guimei Xing; Lianwei Qu collected and identified the plant material; Guimei Xing assembled and annotated the cp genome and manuscript preparation; Huihua Zhang, Yanqiu Zhang, and Jiaojiao Lu contributed significantly to phylogenetic analysis; Tianyu Wu and Zengzhi Tian were involved in the interpretation of the data and revised the manuscript critically; and the authors agrees to be accountable for all aspects of the work.
Ethical approval
Ethics statement: The collection of specimen conformed to the requirement of International Ethics, which did not cause damage to the local environment. The process and purpose of this experimental research were in line with the rules and regulations of Liaoning Academy of Agricultural Sciences.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession No. OL350837. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA834607, SRR19070076, and SAMN28052728, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Chen F, Liu T, Zhou LL. 2001. Research on the biological and germinating characters of wild Tulipa. J Shihezi Univ. 5(3):197–200.
- Diana E. 2013. The genus Tulipa. Kew (UK): Royal Botanic Gardens.
- Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Han B, Zhang K, Huang L. 2014. Amana wanzhensis (Liliaceae), a new species from Anhui, China. Phytotaxa. 177(2):118–124.
- Hoog MH. 1973. On the origin of Tulipa, lilies and other Liliaceae. London: Royal Horticulture Society; p. 47–64.
- Jiao F, Liu Q, Sun GF, Lin QW, Li XD, Zhang JZ. 2015. Study on the germination traits of two kinds of wild tulip seeds in Sinkiang. North Hortic. 2:55–60.
- Ju X, Shi G, Hou Z, Wu C, Liu G, Cao C, Tang N. 2020. Characterization of the complete chloroplast genome of Tulipa iliensis (Liliaceae). Mitochondrial DNA B Resour. 5(3):2362–2363.
- Ju X, Tang N, Shi G, Ye R, Hou Z. 2020. Complete chloroplast genome of Tulipa buhseana (Liliaceae). Mitochondrial DNA B Resour. 5(3):2360–2361.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Mei LJ, Tan DY. 2006. Phenological characteristics in different populations of Tulipa iliensis Regel and Tulipa altaica Pall ex Spreng. J Xinjiang Agric Univ. 29(4):18–21.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(1):6–73.
- Stork AL. 1984. Tulipes sauvages et cultivées. Genève: Série documentaire 13 des Conservatoire et Jardin botaniques.
- Tang J, Wang FZ. 1980. The Flora of China. Vol. 14. Beijing: Science Publishing House; p. 81–97.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14(5):1024–1031.
- Zhou JL, Yin PP, Yang C, Zhao YP. 2019. The complete chloroplast genome of Tulipa altaica (Liliaceae), a wild relative of tulip. Mitochondrial DNA B Resour. 4(1):2017–2018.