Abstract
Cynoglossum amabile Stapf & J. R. Drumm., 1906 is a traditional Chinese herbal medicine from southwest China. To better determine its phylogenetic relatedness to other Boraginaceae species, the chloroplast (cp) genome of C. amabile was sequenced. The complete cp genome of C. amabile is 151,532 bp in length, containing a small single-copy (SSC) region with a length of 17,366 bp, a large single-copy (LSC) region with a length of 82,902 bp, and a pair of inverted repeats (IRs) regions each with a length of 25,632 bp. The overall GC content of the cp genome is 37.4%. The maximum-likelihood phylogenetic tree showed that Bothriospermum zeylanicum (J. Jacq.) Druce, 1917 was closely related to C. amabile.
Introduction
Cynoglossum amabile Stapf & J. R. Drumm., 1906 is a perennial herb within the family Boraginaceae, and is widely distributed in the mountainous areas of southwest China (). It grows in hillside meadows, alpine shrubs, dry roads, or coniferous forest edges at an altitude of 1250–4565 meters (Zhang et al. Citation2014). This species is one of the most common ethnic medicinal materials in the producing area. Local people often use it to treat cough, vomiting, fractures, dysentery, and other diseases (Li and Zhao Citation2010). Studies have shown that C. amabile contains a variety of alkaloids, of which the main components are pyrrolizidine alkaloids with high clinical value. In addition, extracts of C. amabile were shown to have inhibitory effects on certain bacteria and fungi (Wang et al. Citation2014), indicating the plant’s high medicinal potential. To date, its chloroplast (cp) genome has yet to be characterized. cp genomes are valuable tools for determining species identity (Li et al. Citation2015), phylogenetic relationships, and germplasm diversity. Here, we reported the complete cp genome sequence of C. amabile, aiming to provide a useful resource for the phylogenetic studies of Boraginaceae.
Materials and methods
DNA extraction and sequencing
The fresh leaves of C. amabile were collected from Nanxi Village, Huangshan Town, Yulong County, Yunnan Province, China (coordinates: 100°8′59.93″E, 26°46′8.02″N; altitude: 3103 m). The collection of plant materials was in accordance with local regulations under the permission of local authorities. The voucher specimen (SWFU20210756MFY) was deposited at the herbarium of Southwest Forestry University, China (http://bbg.swfu.edu.cn/, Yu Xiao, email: [email protected]). The total genomic DNA was extracted from dried plant leaf specimens using the CTAB extraction method (Doyle and Doyle Citation1987). The paired-end sequencing reads were generated by the Illumina HiSeq 2500 sequencing platform (Illumina, San Diego, CA). After removing the poor-quality reads, 3.38 Gb clean data were obtained. The complete cp genome was assembled using NOVOPlasty with the following parameters: wordize = 102; base coverage = 171.44; and k = 75, 85, 95, 105, 115, and 127 (Dierckxsens et al. Citation2017). The complete cp genome of C. amabile was a typical quadripartite structure (Figs. S1 and S2). The cp genome was annotated in Geneious Prime ver. 2021.2.2 (Kearse et al. Citation2012) using Trigonotis peduncularis (MZ911745.1) as the reference sequence. The cp genome sequence of C. amabile has been submitted to GenBank under the accession number NC_061706. The cpgview (http://www.1kmpg.cn/cpgview) was used to draw the physical map of the cp genome.
Simple sequence repeat analysis
The presence of SSR loci in the cp genome of C. amabile was analyzed using the online detection tool MISA-web (https://webblast.ipk-gater-sleben.de/misa/) (Beier et al. Citation2017). The minimum number of repeats was set to 10 for mononucleotides; five for dinucleotides; and four for trinucleotides, tetranucleotides, and pentanucleotides.
Phylogenetic analysis
A phylogenetic tree was reconstructed to ascertain the phylogenetic relationship of C. amabile and 13 other species of Boraginaceae, with Agastache rugosa and Ajuga forrestii as outgroup taxa (Table S1). The MAFFT ver. 7 program (scoring matrix = 200, PAM k = 2, gap open penalty = 1.53, offset value = 0.123) was used for multiple alignment between the cp genome sequences of these 16 taxa, and the sequence differences were identified (Katoh and Standley Citation2013). Then, the alignment results were checked using Geneious Prime with the output file in the format of *.net. A maximum-likelihood (ML) tree was constructed with RAxML ver. 8.0.0 (m = GTR + GAMMA, bootstrap = 1000) (Stamatakis Citation2014) and was visualized with FigTree ver. 1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
Results and discussion
Structural characteristics
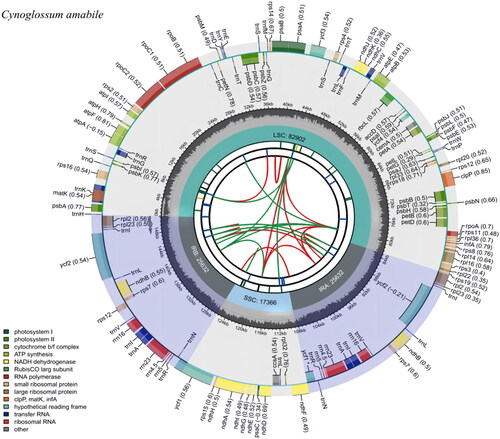
The complete cp genome of C. amabile is 151,532 bp in length with the typical double-stranded circular tetrad structure (), containing a small single-copy (SSC) region with a length of 17,366 bp, a large single-copy (LSC) region with a length of 82,902 bp, and a pair of inverted repeats (IRs) regions with a length of 25,632 bp each. The contents of A, T, C, and G are 31.0%, 31.6%, 19.0%, and 18.4%, respectively. The overall GC content of the whole genome is 37.4% (LSC, 35.19%; SSC, 30.86%; IR, 43.09%). In total, 130 genes were annotated, including 85 protein-coding genes (PCGs), eight ribosomal RNA genes (rRNAs), and 37 transfer RNA genes (tRNAs). Of the 130 genes encoded, there are 17 dual-copy genes, including two ribosome macrounit genes (rpl2, rpl23), one ribosome subunit gene (rps7), one NADH dehydrogenase gene (ndhB), four rRNA genes (rrn5, rrn4.5, rrn23, rrn16), seven tRNA genes (trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, trnV-GAC, and trnA-UGC), and two functionally unknown genes (ycf2 and ycf1).
Figure 2. Gene map of Cynoglossum amabile chloroplast genome. The figure consists of several circles, and the information about each circle from the center to outward is as follows: the circle nearest to the center shows the forward and reverse repeats by red and green arcs, respectively. The second and third circles indicate the tandem repeats and microsatellite sequences by short bars, respectively. The fourth circle shows the position of the LSC, SSC, IRA, and IRB regions, respectively. The fifth circle shows the GC content. The outer circle indicates the gene functions. Different colors are used to show different functional categories, as shown in the bottom left of the figure.
Sequence repeat analysis
A total of 30 SSRs were detected by the online software MISA-web (Table S2), with the numbers of mono-, di-, tri-, tetra-, and pentanucleotides SSRs being 20, 6, 1, 0, and 0, respectively. The number of mononucleotides is the highest, consistent with a previous study in which the mononucleotide had the highest number of occurrences of A and T (Yang et al. Citation2021). Our result is also consistent with the previous report that the mononucleotide repeats of the cp genome are biased toward A/T repeats (Kuang et al. Citation2011). Dinucleotide repeats have AT/TA with 4 and 2 occurrences, respectively; trinucleotide has only TTC. Previous studies have suggested that mononucleotides in the cp genome are more likely to exhibit intraspecific variation if located in a noncoding single-copy region (Ebert and Peakall Citation2009), and analysis of SSRs revealed that 16 mononucleotides were distributed in the noncoding single-copy region. The cp genome of C. amabile is enriched in AT, suggesting that it is easier to unpack because AT binding has one less hydrogen bond than GC binding, and the energy required to break AT binding is much lower than that of GC bonds. Therefore, AT repeat motifs are more likely to be included in SSR repeat motifs in the cp genome (Qiao et al. Citation2019; Zhao et al. Citation2021).
Phylogenetic analyses
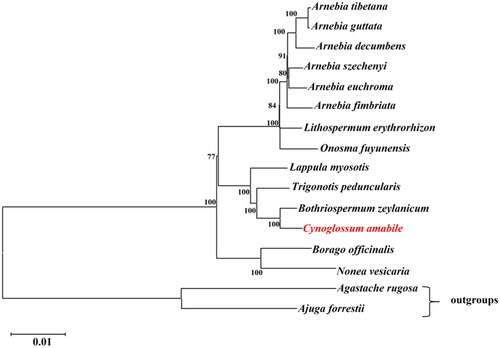
The phylogenetic analysis revealed that all species of Boraginaceae formed one monophyletic clade. The phylogenetic tree indicated that Bothriospermum zeylanicum (J. Jacq.) Druce, 1917 was a sister taxon to C. amabile (). Our cp genomic data of C. amabile contributes to the growing quantity of cp genomes for phylogenetic and evolutionary studies in the family Boraginaceae.
Figure 3. ML phylogenetic tree based on complete chloroplast genome of C. amabile and 15 other species. Numbers in the nodes are the bootstrap values from 1000 replicates. The following sequences were used: Arnebia decumbens ON529954 (Sun et al. Citation2022), Arnebia euchroma ON529958, Arnebia fimbriata ON529943, Arnebia guttata ON529956, Arnebia szechenyi ON529949, Arnebia tibetana MT975392 (Park et al. Citation2020), Borago officinalis NC_046796 (Guo et al. Citation2020), Bothriospermum zeylanicum NC_065834, Cynoglossum amabile NC_061706, Lappula myosotis NC_060614, Lithospermum erythrorhizon MT975394, Nonea vesicaria OL335187 (Carvalho Leonardo et al. Citation2022), Onosma fuyunensis NC_049569 (He et al. Citation2021), Trigonotis peduncularis MZ911745 (Wu et al. Citation2022), Agastache rugosa MW760849 (Wang et al. Citation2021), and Ajuga forrestii NC_048512 (Tao et al. Citation2019).
Conclusions
In this study, the cp genome sequence of C. amabile was assembled, annotated, and analyzed. B. zeylanicum was found to be closely related to C. amabile. These results provide the basis for the molecular identification of Cynoglossum plants. This study opens up new avenues for future research and obtains the genomic information of the cps of Boraginaceae taxa on this basis.
Author contributions
X. Y. collected the molecular materials and conceived the study; Z.-N. Z. drafted the manuscript and analyzed the experimental data; X. Y. revised the manuscript. Both authors provided comments and final approval.
Supplemental Material
Download MS Word (452.1 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession number NC_061706. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA797868, SRR17640219, and SAMN25038855, respectively.
Additional information
Funding
References
- Beier S, Thiel T, Münch T, Scholz U, Mascher M. 2017. MISA-web: a web server for microsatellite prediction. Bioinformatics. 33(16):2583–2585.
- Carvalho Leonardo I, Barreto Crespo MT, Capelo J, Bustos Gaspar F. 2022. The complete plastome of Nonea vesicaria (L.) Rchb. (Boraginaceae), the first chloroplast genome belonging to the Nonea genus. Mitochondrial DNA B Resour. 7(7):1302–1304.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Ebert D, Peakall ROD. 2009. Chloroplast simple sequence repeats (cpSSRs): technical resources and recommendations for expanding cpSSR discovery and applications to a wide array of plant species. Mol Ecol Resour. 9(3):673–690.
- Guo X, Wang X, Wang Q, Liu C, Zhang R, Cheng A, Sun J. 2020. The complete chloroplast genome sequence of Borago officinalis Linn. (Boraginaceae) and its phylogenetic analysis. Mitochondrial DNA B Resour. 5(2):1461–1462.
- He Y, Xu X, Liu Q. 2021. The complete chloroplast genome of Onosma fuyunensis Y. He & QR Liu and its phylogenetic analysis. Mitochondrial DNA B Resour. 6(11):3142–3143.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kuang DY, Wu H, Wang YL, Gao LM, Zhang SZ, Lu L. 2011. Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): implication for DNA barcoding and population genetics. Genome. 54(8):663–673.
- Li W, Zhao FC. 2010. Research on quality specification of herb Cynoglossum officinale. J Xinjiang Med Univ. 33(9):1036–1038.
- Li X, Yang Y, Henry RJ, Rossetto M, Wang Y, Chen S. 2015. Plant DNA barcoding: from gene to genome. Biol Rev Camb Philos Soc. 90(1):157–166.
- Park I, Yang S, Song JH, Moon BC. 2020. Dissection for floral micromorphology and plastid genome of valuable medicinal borages Arnebia and Lithospermum (Boraginaceae). Front Plant Sci. 11:606463.
- Qiao YG, He JX, Wang YF, Cao YP, Jia MJ, Zhang XR, Liang JP, Song Y. 2019. Analysis of chloroplast genome and its characteristics of medicinal plant Sophora flavescens. Acta Pharm Sin. 54(11):2106–2112.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Sun J, Wang S, Wang Y, Wang R, Liu K, Li E, Qiao P, Shi L, Dong W, Huang L, et al. 2022. Phylogenomics and genetic diversity of Arnebiae Radix and its allies (Arnebia, Boraginaceae) in China. Front Plant Sci. 13:920826.
- Tao AE, Zhao FY, Xia CL. 2019. Characterization of the complete chloroplast genome of Ajuga forrestii (Lamiaceae), a medicinal plant in southwest of China. Mitochondrial DNA B Resour. 4(2):3969–3970.
- Wang F, Liang Q, Yang JZ, Wu JR. 2014. Anti-microbial activity of the extracts from Cynoglossum amabile. J Northwest Forest Univ. 29(5):125–128 + 175.
- Wang Y, Wang H, Zhou B, Yue Z. 2021. The complete chloroplast genomes of Lycopus lucidus and Agastache rugosa, two herbal species in tribe Mentheae of Lamiaceae family. Mitochondrial DNA B Resour. 6(1):89–90.
- Wu JH, Li HM, Lei JM, Liang ZR. 2022. The complete chloroplast genome sequence of Trigonotis peduncularis (Boraginaceae). Mitochondrial DNA B Resour. 7(3):456–457.
- Yang H, Wang L, Chen H, Jiang M, Wu W, Liu S, Wang J, Liu C. 2021. Phylogenetic analysis and development of molecular markers for five medicinal Alpinia species based on complete plastome sequences. BMC Plant Biol. 21(1):1–16.
- Zhang XJ, Chen LL, Hao JJ. 2014. Resource survey of Cynoglossum amabile Stapf et Drumm and the same genus plants. Chin J Ethnomed Ethnopharm. 23(15):11–16.
- Zhao Q, Yu JX, Qin YW, Ren XY, Gao HD, Wu ZG, Jiang CX. 2021. Assembly and sequence analysis of chloroplast genome of Championella sarcorrhiza based on high-throughput sequencing. Chin Tradit Herb Drugs. 52(6):1744–1750.

