Abstract
Despite having many historically reported ethnomedicinal uses, Centaurium erythraea Rafn (Rafn and Buchs, 1800; common centaury) also produces cytotoxic secondary metabolites, and its presence should be carefully monitored. In this study, the complete chloroplast of Centaurium erythraea subsp. majus (Hoffmanns. & Link) M.Laínz (Laínz, 1971) isolate BPTPS121 is described, being the first available plastome belonging to the Centaurium genus. The chloroplast genome (GenBank accession number: ON641347) is 153,107 bp in length with 37.9% GC content, displaying a quadripartite structure that contains a pair of inverted repeat regions (25,166 bp each), separated by a large single-copy (84,388 bp) and small single-copy (18,387 bp) regions. A total of 129 genes were predicted, including 37 tRNA genes, eight rRNA genes, and 84 protein-coding genes. The phylogenetic analysis showed that isolate BPTPS121 is placed under the Gentianaceae family, belonging to the Gentianales order. The maximum-likelihood tree supports the already described lineage divergence in the Gentianaceae family, with C. erythraea subsp. majus belonging to the Chironieae tribe positioned below the Exaceae tribe and above the Potalieae and the entire Gentianeae tribes. This study will contribute to conservation, phylogenetic, and evolutionary studies, as well as DNA barcoding applications for food, feed, and supplements safety purposes.
Centaurium Hill (Hill et al. Citation1756), a genus of flowering plants in the Gentianaceae family commonly known as ‘centauries’, has included ca. 108 species in the past. This previously polyphyletic genus has since been reclassified and redistributed among different genera. For example, ca. 25 species of New World centauries were transferred to the Zeltnera G.Mans. genus, ca. five species to Gyrandra Griseb. (Mexico and Central America), and ca. five to Schenkia Griseb. (Australia and Pacific) (Mansion Citation2004). This redistribution has left Centaurium sensu stricto with ca. 20 species (Struwe Citation2014), distributed mainly around the Western Mediterranean region while reaching the Balkan Peninsula.
Centaurium erythraea Rafn (Rafn and Buchs Citation1800; common centaury) is the most abundant species of which many ethnomedicinal uses have been historically reported. Significantly, it is one of the plants described and depicted in the work De Materia Medica by the celebrated Greek medical writer Dioscorides (40–90 BCE). Common putative uses have been described as anti-bacterial, anti-fungal, anti-leishmanial, insecticidal, anti-oxidant, anti-inflammatory, anti-diabetic, and anti-proliferative, as well as gastroprotective, hepatoprotective, dermoprotective, and neuroprotective, among others (El Menyiy et al. Citation2021). These various pharmacological properties arise from the production of several classes of secondary metabolites, namely xanthonoids, terpenoids, flavonoids, phenolic acids, and fatty acids. From these, significant antibacterial activity has been attributed to two secoiridoid glycosides, swertiamarin and sweroside, as well as cytotoxicity (Kumarasamy et al. Citation2003) and, as such, their presence should be carefully monitored.
The material of Centaurium erythraea subsp. majus (Hoffmanns. & Link) M.Laínz (Laínz Citation1971) analyzed (BioSample: SAMN28118559; ) was collected from a wild population in Torres Vedras municipality (Dois Portos) in Portugal (collection date: 2020-06-15; location: 39.06612 N, 9.1788 W). This plant material was identified as isolate BPTPS121 with a specimen being conserved at the LISE Herbarium (INIAV, Oeiras, Portugal; Jorge Capelo: [email protected]) under the voucher LISE: 96442 (identified by: Jorge Capelo).
Figure 1. Detail of C. erythraea subsp. majus isolate BPTPS121 (BioSample: SAMN28118559) after being dried and before being mounted and conserved at the LISE Herbarium (INIAV, Oeiras, Portugal; Jorge Capelo: [email protected]) under the voucher LISE: 96442 (identified by: Jorge Capelo). This isolate was collected from a wild population in Torres Vedras municipality (Dois Portos) in Portugal (collection date: 15 June 2020; location: 39.06612 N, 9.1788 W).

Immediately after collection, young leaves from the selected specimen were frozen in liquid nitrogen and stored at −80 °C until further processing. Total genomic DNA was extracted from the preserved material using an adaptation of the Doyle and Doyle (Citation1987) methodology. After quantity (Qubit 4 Fluorometer, Thermo Fisher Scientific, Waltham, MA) and quality (NanoDrop ND-1000, Thermo Fisher Scientific, Waltham, MA) evaluation, the obtained DNA was sent to Genoscope (Évry, France) for sequencing. DNA was first sonicated using the Covaris E210 Focused Ultrasonicator instrument (Woburn, MA), and then libraries were prepared with the NEBNext Ultra II DNA Library Prep Kit (New England Biolabs, Ipswich, MA). Finally, sequencing was performed using 151 base-length read chemistry in a paired-end flow cell on the Illumina NovaSeq 6000 sequencing platform (San Diego, CA).
The about 40 million high-quality paired-end reads obtained (SRA: ERR10047930) were used to assemble the complete chloroplast genome (sequence coverage: 972×) using the GetOrganelle pipeline (v1.7.3.1) (Jin et al. Citation2020). The pipeline was used following the typical recipe suggested for Embryophyta plant plastome assembly (https://github.com/Kinggerm/GetOrganelle) while setting the flags ‘--max-reads’ and ‘--reduce-reads-for-coverage’ to 25 million and one thousand, respectively (see Supplemental material for additional details). The plastome annotation was performed using the GeSeq tool (Tillich et al. Citation2017) using the default parameters and the provided 3rd party stand-alone annotators Chloë (v0.1.0). A subsequent manual curation of the obtained annotations was performed using Geneious Prime 2022.0.1 (https://www.geneious.com) to compare them with the results obtained by performing a BLAT search on GeSeq (protein, rRNA, tRNA, DNA search identities set to 90%; see Supplemental material for additional details).
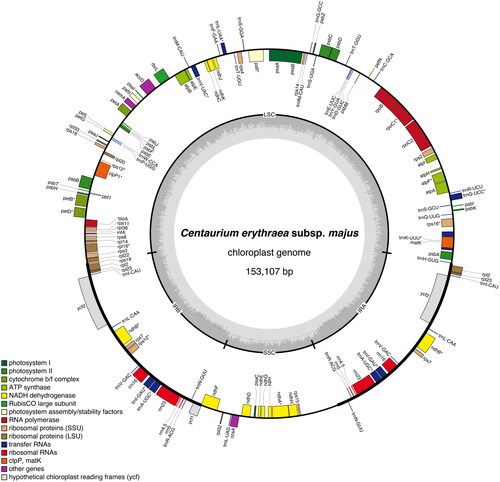
The chloroplast genome of C. erythraea subsp. majus isolate BPTPS121 (GenBank accession number: ON641347; ) is 153,107 bp in length with 37.9% GC content, displaying a quadripartite structure that contains a pair of inverted repeat (IR) regions (25,166 bp; GC content 43.4%), separated by a large single-copy (LSC) region (84,388 bp; GC content 36.0%) and a small single-copy (SSC) region (18,387 bp; GC content 31.7%). A total of 129 genes were predicted (113 of them unique), including 37 tRNA genes (30 of them unique), eight rRNA genes (four of them unique), and 84 protein-coding genes (79 of them unique).
Figure 2. Graphical map of the complete chloroplast genome of Centaurium erythraea subsp. majus isolate BPTPS121 based on the conversion of annotations openly available in GenBank (accession number: ON641347), color coded based on their functional group, using OrganellarGenomeDRAW (OGDRAW) version 1.3.1 (Greiner et al. Citation2019). Genes inside the circle are transcribed clockwise, genes outside the circle counterclockwise, and intron-containing genes are marked by an asterisk (*). LSC: large single-copy region; SSC: small single-copy region; IRA, IRB: inverted repeats (IR). The dark grey inner ring represents the GC content, while the complementary light grey ring represents the AT content.
The phylogenetic analysis (see Supplemental material for additional details) was performed using the concatenated nucleotide sequences coding for the shared proteome (65 coding sequences) extracted from a selected dataset. The dataset was composed of 17 verified and complete chloroplast genomes belonging to the Gentianaceae family available in GenBank (accession date: 26 June 2022), with only one species representing each genus. The selected dataset also included the complete chloroplast genome of C. erythraea subsp. majus isolate BPTPS121 obtained in this study and three additional sequences used as outgroups in the phylogenetic analysis: Catharanthus roseus (L.) G.Don (NC_021423; Apocynaceae family) belonging to the Gentianales order but not from the Gentianaceae family, Nonea vesicaria (L.) Rchb. (NC_060826; Boraginales order) belonging to the same lamiids clade but from a different order, and Tilia platyphyllos Scop. (NC_062378; Malvales order) from the malvids clade. The sequences were aligned using MAFFT v7.450 (Katoh and Standley Citation2013) and further analyzed with the IQ-TREE 2 software package (Minh et al. Citation2020). The best-fit substitution model (TVM + F+I + I+R2 chosen according to the Bayesian information criterion) was selected according to ModelFinder (Kalyaanamoorthy et al. Citation2017), followed by a tree reconstruction () using IQ-TREE (Nguyen et al. Citation2015) using ultrafast bootstrap with UFBoot (10,000 replicates) (Hoang et al. Citation2018).
Figure 3. Maximum-likelihood tree inferred from the sequences coding for the shared proteome from Centaurium erythraea subsp. majus isolate BPTPS121 and 17 verified and complete chloroplast genomes belonging to the Gentianaceae family available in GenBank (accession date: 26 June 2022). The numbers attached to the branches show the SH-aLRT and the UFBoot2 percent supports (SH-aLRT/UFBoot2). Catharanthus roseus (L.) G.Don (NC_021423; lamiids clade, Apocynaceae family), Nonea vesicaria (L.) Rchb. (NC_060826; lamiids clade, Boraginales order), and Tilia platyphyllos Scop. (NC_062378; malvids clade, Malvales order) were used as the outgroups.
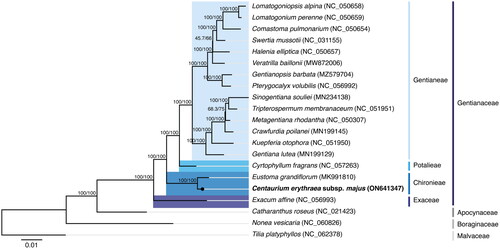
The maximum-likelihood tree showed that C. erythraea subsp. majus isolate BPTPS121 is placed under the Gentianaceae family, belonging to the Gentianales order. The Gentianaceae family has six tribes in the current classification: the Chironieae, Exaceae, Gentianeae, Helieae, Saccifolieae, and Potalieae (Struwe Citation2014). These tribes are unevenly represented in GenBank’s genome resources, with Gentianeae having 163 unique, verified, and complete chloroplast genomes, Chironieae, Exaceae, and Potalieae with one unique entry each, and Helieae and Saccifolieae with none. Using the available data, the phylogenetic analysis performed supports that the Chironieae tribe (C. erythraea subsp. majus isolate BPTPS121 (ON641347) and Eustoma grandiflorum (MK991810)) is the second most basally positioned tribe, below Exaceae (Exacum affine; NC_056993), and above Potalieae (Cyrtophyllum fragrans; NC_057263) and the entire Gentianeae tribe, with 100/100 percent support (SH-aLRT/UFBoot2). This tree, therefore, supports the already described lineage divergence in the Gentianaceae family (Struwe Citation2014), with complete chloroplast genomes of isolates from the Helieae and Saccifolieae tribes still missing in the databases, as well as for the still uncertain placement of Voyria. The phylogenetic analysis performed using the concatenated amino acid sequences of the shared proteomes also supports the same phylogenetic result (see Supplemental material for additional details).
This study describes the chloroplast genome of C. erythraea subsp. majus isolate BPTPS121, the first described plastome belonging to the Centaurium genus. This complete genome will contribute to conservation, phylogenetic, and evolutionary studies in the Gentianaceae family. It will also support DNA barcoding applications for food, feed, and supplements safety and quality purposes that target detecting species that produce secondary metabolites with cytotoxic potential.
Ethical approval
No ethical approval was required to collect and study the specimen described in this manuscript. The species is not under legal protection status, either by national or European Union legislation, namely the 92/43/CEE Directive. A careful nondestructive collection protocol for voucher sampling was followed to guarantee the full future reproductive viability of the studied plant population.
Author contributions
The authors had the following contribution to the paper: MTBC and FBG – conception and design; FBG – analysis and interpretation of the data; JC – collection and taxonomic identification of the studied specimen; ICL – sample processing; AA – shotgun library preparation and sequencing; FD – data management and submission; ICL, AA, FD, JC, MTBC, and FBG – manuscript drafting, critical revision for intellectual content, and final approval of the version to be published. All authors agree to be accountable for all aspects of the work herein presented.
Disclosure statement
No potential competing or conflict of interests was reported by the authors.
Data availability statement
The data that support this study is openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession number ON641347. The associated BioProject, BioSample, and SRA numbers are PRJNA848681, SAMN28118559, and ERR10047930, respectively.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- El Menyiy N, Guaouguaou F-E, El Baaboua A, El Omari N, Taha D, Salhi N, Shariati MA, Aanniz T, Benali T, Zengin G, et al. 2021. Phytochemical properties, biological activities and medicinal use of Centaurium erythraea Rafn. J Ethnopharmacol. 276:114171.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Research, 47(W1), W59–W64. doi:10.1093/nar/gkz238
- Hill J, Benning R, Boyce S, Burgess T, Collins B, Grignion C, Newbery J, Roberts H, Smith A, Smith J, et al. 1756. The British herbal: an history of plants and trees, natives Britain, cultivated for use, or raised for beauty. For T. Osborne and J. Shipton [etc.]. London.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the Ultrafast Bootstrap Approximation. Mol Biol Evol. 35(2):518–522.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley D. M. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumarasamy Y, Nahar L, Cox PJ, Jaspars M, Sarker SD. 2003. Bioactivity of secoiridoid glycosides from Centaurium erythraea. Phytomedicine. 10(4):344–347.
- Laínz M. 1971. Aportaciones Al Conocimiento De La Flora Gallega. Vol. VII. Madrid: Univers. Laboral de Gijón, Inst. Forestal de Investigaciones y Experiencias.
- Mansion G. 2004. A new classification of the polyphyletic genus Centaurium Hill (Chironiinae, Gentianaceae): description of the New World endemic Zeltnera, and reinstatement of Gyrandra Griseb. and Schenkia Griseb. Taxon. 53(3):719–740.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Rafn CG, Buchs CL. 1800. Danmarks og Holsteens flora. Vol. 2. Trykt paa boghandler C.L. Buchs Forlag, hos Sebastian Popp.
- Struwe L. 2014. Classification and evolution of the family Gentianaceae. In: Rybczyński JJ, Davey MR, Mikuła A, editors. The Gentianaceae—Volume 1: characterization and ecology. Berlin, Heidelberg: Springer; p. 13–35.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
