Abstract
Cheilinus trilobatus Lacépède, 1801 is a species of genus Cheilinus. In this study, we sequenced the complete mitochondrion genome of C. trilobatus. The mitochondrial genome was 17,292 bp, consisting of 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and one non-coding control region (D-loop). The nucleotide composition was 27.31% A, 25.1% T, 17.23% G, and 30.36% C. Phylogenetic analysis suggested that C. trilobatus was closely related to Cheilinus oxycephalus. The complete mitogenome of C. trilobatus provided basic data for the genetic diversity conservation of this species.
1. Introduction
Cheilinus trilobatus Lacépède, 1801 is a common species belonging to the family Labridae. It usually inhabits in shallow reefs, especially in areas with rich coral and algal cover (Khalaf and Disi Citation1997). It feeds on mollusks, crustaceans, and sea urchins, and occasionally takes fishes (Myers Citation1991). Previous studies on this species have focused on its chromosome analysis and levels of heavy metals. However, the study about the molecular biological properties of C. trilobatus has not been published. In this study, we first sequenced the complete mitochondrial genome of C. trilobatus, and performed a phylogenetic analysis among Cheilinus with the available mitogenomic sequences. It may provide some basic data for the genetic diversity conservation of this species.
2. Materials
Cheilinus trilobatus was obtained from the Qilianyu (16°55′–17°00′N, 112°12′–112°21′E), Xisha Islands, China in April 2020 (). All specimens were deposited at South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences ([email protected], Voucher specimen: SYCY20210420001).
3. Methods
Genomic DNA was extracted from an adult’s dorsal muscle using E.Z.N.A.® Tissue DNA Kit (OMEGA, China) following the manufacturer’s instructions. DNA library preparation and 150-bp paired-end sequencing were performed on the Illumina HiSeq platform. Raw reads used for assembling the mitochondrial genome were deposited into the NCBI SRA (SRA accession: SRR18531815). The raw data was assembled by using GetOrganelle version 1.7.5.3 (Jin et al. Citation2020). The complete mitochondrial of C. trilobatus was annotated by MITOS2 web server (Bernt et al. Citation2013). Protein-coding genes (PCGs) and rRNAs were annotated by comparing with the complete mitochondrial genome of the Cheilinus undulatus Rüppell, 1835 (Matthew et al. Citation2018). The 13 common protein-coding genes in each complete mitochondrial genome of 12 species were aligned with the genes in C. trilobatus using MAFFT 7.037 (Katoh and Standley Citation2016). Then, model-finder var 1.6 was run to select the best-fit model and the mtVer + F + R3 model was chosen. Finally, iqtree 2.0 was used to construct a phylogenetic tree with 1,000 bootstraps based on the ML method.
4. Results
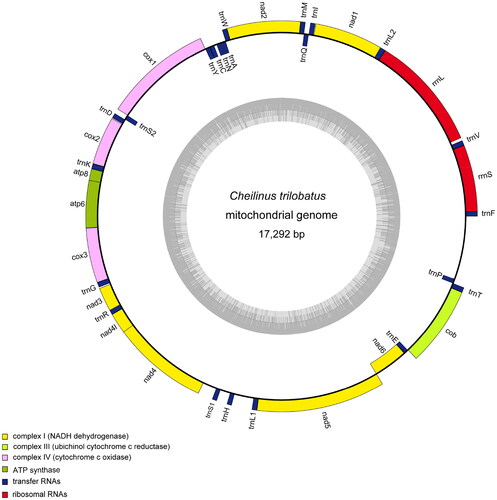
The annotated complete mitochondrial genome sequence was submitted to NCBI (GenBank: OM994969). It was 17,292 bp in length containing 13 PCGs (Cyt b, ATP6, ATP8, COX1-3, ND1-6, ND4L), 22 tRNA genes, two rRNA genes (12S and 16S rRNA), and one control region (CR or D-Loop) (). Eight tRNA genes (Gln, Ala, Asn, Cys, Tyr, Ser, Glu, and Pro) and NADH dehydrogenase subunit 6 (ND6) are encoded on the light strand (L-strand), the other 29 genes are encoded on the heavy strand (H-strand). The nucleotide composition was 27.31% A, 25.1% T, 17.23% G, and 30.36% C. Almost all of 13 PCGs for C. trilobatus share the regular initiation codon ATG except COI gene with GTG, and ND3 gene with GTT. There are three different patterns of termination codons: eight PCGs terminated with the stop codons TAA or TAG, whereas five PCGs (COX2, COX3, ND3, ND4, Cyt b) end with incomplete form TA − or T–.
Figure 2. Mitochondrial genome map of Cheilinus trilobatus. The circular map of C. trilobatus was drawn using the OGDRAW program. The map consists of two circles and information about each circle is as follows: the inner circle indicates the GC content, and the external circle indicates the genes having different colors based on their functions.
5. Discussion and conclusion
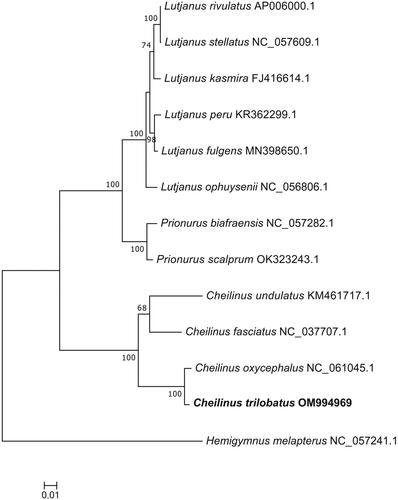
To further investigate the phylogenetic relationships of Cheilinus and the position of C. trilobatus, phylogenetic trees were constructed based on the whole mitochondrial genome (). The phylogenetic result indicated that C. trilobatus was closely related to Cheilinus oxycephalus than C. undulatus and Cheilinus fasciatus, which was congruent with the result from the barcode of life data system (Ward et al. Citation2009). This study provides an important molecular resource for further study on the genus Cheilinus.
Figure 3. Maximum-likelihood (ML) phylogenetic tree based on complete mitogenome sequences. Numbers near the nodes represent ML bootstrap values. The following sequences were used: Cheilinus oxycephalus NC_061045.1, Cheilinus fasciatus NC_037707.1, Cheilinus undulatus KM461717.1, Lutjanus stellatus NC_057609.1, Lutjanus rivulatus AP006000.1, Lutjanus kasmira FJ416614.1, Lutjanus fulgens MN398650.1, Lutjanus peru KR362299.1, Lutjanus ophuysenii NC_056806.1, Prionurus biafraensis NC_057282.1, Prionurus scalprum OK323243.1, and Hemigymnus melapterus NC_057241.1.
Ethical approval
The data collection of fishes was carried out with the permission of related institution, and complied with national or international guidelines and legislation.
Author contributions
C.H. Li designed and conceived this work. T. Wang, Y.P. Li, Y. Liu, Y.Y. Xiao, P. Wu, and L. Lin collected the samples. Q. Ma conducted DNA experiment and analyzed the data. All authors read, revised, and approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession number OM994969. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA820490, SRR18531815, and SAMN27010482, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):1–31.
- Khalaf MA, Disi AM. 1997. Fishes of the Gulf of Aqaba. Publication of the Marine Science Station, No. 8.
- Katoh K, Standley DM. 2016. A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics. 32(13):1933–1942.
- Matthew P, Manjaji-Matsumoto BM, Rodrigues KF. 2018. Complete mitochondrial genome of six Cheilinus undulatus (Napoleon Wrasse): an endangered marine fish species from Sabah, Malaysia. Mitochondrial DNA B Resour. 3(2):943–944.
- Myers RF. 1991. Micronesian reef fishes. 2nd ed. Guam: Coral Graphics. p. 298.
- Ward RD, Hanner R, Hebert PDN. 2009. The campaign to DNA barcode all fishes. J Fish Biol. 74(2):329–356.

