Abstract
The complete mitochondrial genome of Brachyrhynchus triangulus Bergroth, 1889 was sequenced and annotated in the present study. It was a typical circular DNA molecule of 15,170 bp, with 37 genes and a control region. The percentages of A, C, G, and T nucleotides in the genome of B. triangulus were 41.1%, 17.4%, 11.9%, and 29.4%, respectively. Thirteen protein-coding genes (PCGs) start with a ATN codon or a TTG codon and terminate with a TAA codon or a TAG codon or a single T residue. With the exception of tRNASer(AGN), each of the 22 tRNA genes had a clover-leaf structure and ranged in length from 62 to 69 bp. The length of lrRNA and srRNA was 1241 bp and 828 bp, respectively. The control region had a length of 708 bp and an A + T content of 74.6%. The sister relationship between B. triangulus and Brachyrhynchus hsiaoi is supported by the phylogenetic tree. Additionally, it proved the sister relationship between Mezirinae and Aneurinae, supporting the classical taxonomy of the Aradidae.
Brachyrhynchus triangulus (Hemiptera: Aradidae) was discovered in Burma by Bergroth (Bergroth Citation1889). Aradids were known as flat bugs because of their strongly flattened dorso-ventrals and bark bugs because they live under the bark of rotting trees. Flat bugs are brown to black in color (Bhagat Citation2015). They are mysterious and their economic functions have not been discovered (Chandra and Kushwaha Citation2015). In this study, we first present the complete mitogenome of B. triangulus and provide valuable genetic information for further research on the classification and genomics of Aradidae.
The specimens used in this experiment were obtained on 26 March 2020, from the Tongbiguan Nature Reserve, Yingjiang, Yunnan province (24.70579N, 97.93179E), and were identified by Professor Xiaoshuan Bai from Inner Mongolia Normal University. The voucher specimen was deposited with the voucher number BtYj-1 in absolute ethanol at −20 °C in the molecular laboratory of Inner Mongolia Normal University (Xiaoshuan Bai: [email protected]). Since Brachyrhynchus triangulus is neither a vertebrate or a protected invertebrate, the reserve gave its consent for the field study to be carried out. High-throughput sequencing techniques were used to extract the mitochondrial genome of B. triangulus, which was then submitted to the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/, accession number: OK323200).
Total DNA was extracted from the feet of B. triangulus using the TIANamp Genomic DNA Kit and sent to BerryGenomics (Beijing, China) for sequencing. Total genomic DNA was extracted using the Illumina NovaSeq 2500 platform. A total of 6 Gb clean data were returned and assembled by IDBA-UD v1.0 (Peng et al. Citation2012), and mitochondrial genome sequence was obtained using a Python software package. The annotation of B. triangulus was done by MITOS (http://mitos.bioinf.uni-leipzig.de/index.py). The secondary structure of tRNA genes was predicted by tRNA scan-SE 2.0 (Lowe and Eddy Citation1997) (http://lowelab.ucsc.edu/tRNAscan-SE/). The 13 PCGs were annotated using a homology alignment method, and we downloaded the nucleotide sequences of 13 PCGs genes of seven species of Aradidae from NCBI. The locations of the 13 PCGs were determined by manual alignment using MEGA7 (Sudhir et al. Citation2016).
The mitochondrial genome of B. triangulus is 15,170 bp and consists of 37 genes. These genes encode 13 protein-coding genes (PCGs), 22 tRNA-coding genes, two ribosomal RNAs, and a control region. Among them, 23 genes encode in the J-strand and 14 genes encode in the N-strand. Except for the rearrangements between tRNAIle and tRNAGln, the sequences and orientations of these genes are identical to the putative ancestral arrangement of Drosophila yakuba (Clary and Wolstenholme Citation1985). This rearrangement is found in seven other sequenced flat bugs, which may be considered a unique feature of the Aradidae mt-genome (Shi et al. Citation2012; Li et al. Citation2016; Song et al. Citation2016). The nucleotide compositions are 41.1% A, 17.4% C, 11.9% G, and 29.4% T, respectively, with a strong AT bias.
The 13 PCGs have a total length of 10,945 bp and can encode a total of 3638 amino acids. In general, the start codon of the mitochondrial gene in invertebrates is commonly ATN, while TTG or GTG has also been found to be the start codon for several insects (Clary and Wolstenholme Citation1985). The complete stop codon is TAA and TAG, and insect mitochondria also have incomplete stop codons, such as TA and T. The incomplete stop codon is usually found between PCG and its adjacent tRNA genes. Ten PCGs are initiated with ATN (six for ATG, three for ATA, and one for ATT) as the start codon, and the others (COI, COII, and ND1) are initiated with TTG. Most PCGs terminate with TAA or TAG as a stop codon, and the others (COII, COIII, ND4, and ND5) terminate with a single T residue.
Except for the dihydrouridine (DHU) arm of tRNASer(AGN), which forms a simple loop, all 22 of the tRNA genes, ranging in length from 62 to 69 bp, have a clover-leaf structure. The overlap between tRNATrp and tRNACys is the largest (8 bp). The length of lrRNA is 1241 bp with 74.3% A + T content, and the length of srRNA is 828 bp with 66.7% A + T content. The 708 bp control region is located between srRNA and tRNAGln, with 74.6% A + T content.
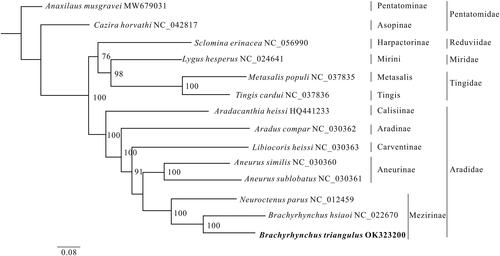
We constructed a phylogenetic tree of B. triangulus using the maximum-likelihood (ML) method to resolve the phylogenetic position of B. triangulus (). The findings indicated a tight relationship between Brachyrhynchus triangulus and Brachyrhynchus hsiaoi. It also demonstrated a sisterly relationship between Mezirinae and Aneurinae, supporting the classical taxonomy of the Aradidae (Heiss and Grebennikov Citation2014; Song et al. Citation2016).
Figure 1. The phylogenetic relationships of the 14 species are derived from analysis of the combined 13 protein-coding sequence data based on the maximum-likelihood (ML) method. The best model is selected using ModelFinder (Kalyaanamoorthy et al. Citation2017) as GTR + F+I + G4, and later the ML tree is inferred from IQ-Tree v.1.6.8 (Lam-Tung et al. Citation2015). The node values are obtained with 1000 repetitions of bootstrap percentages.
Author contributions
Xiaojun Zhu: conceptualization, methodology, formal analysis, resources, investigation, writing – original draft, writing – review and editing, and visualization. Hongge Qian: investigation, writing – review and editing. Xiaoshuan Bai: resources, formal analysis, investigation, writing – review and editing, visualization, supervision, project administration, and funding acquisition.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. OK323200. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA838196, SRR19226807, and SAMN28408660, respectively.
Additional information
Funding
References
- Bergroth E. 1889. Viaggio di Leonardo Fea in Birmania e regioni vicine. XXII. Commentarius de Aradides in Burma et Tenasserim a Fea collectis. Ann Mus Nat Genova. 2(7):736.
- Bhagat RC. 2015. Faunal biodiversity and updated annotated checklist of Pentatomomorpha bugs (Heteroptera: Aradoidea, Coreoidea, Lygeoidea, Pyrrhocoroidea) of Jammu, Kashmir & Ladakh Himalayas (India). Indian J Fundam Appl Life Sci. 5(2):10–17.
- Chandra K, Kushwaha S. 2015. Brachyrhynchus triangulus Bergrot, 1889 (Hemiptera: Aradidae), a new record from India. J Andaman Sci Assoc. 20(2):220–221.
- Clary DO, Wolstenholme DR. 1985. The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 22(3):252–271.
- Heiss E, Grebennikov V. 2014. DNA barcoding of flat bugs (Hemiptera: Aradidae) with phylogenetic implications. Arthropod Syst Phylogeny. 72:213–219.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Lam-Tung N, Schmidt HA, Arndt VH, Quang MB. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Li H, Shi AM, Song F, Cai W. 2016. Complete mitochondrial genome of the flat bug Brachyrhynchus hsiaoi (Hemiptera: Aradidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):14–15.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.
- Peng Y, Leung HCM, Yiu SM, Chin FYL. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 28(11):1420–1428.
- Shi A, Li H, Bai X, Dai X, Chang J, Guilbert E, Cai W. 2012. The complete mitochondrial genome of the flat bug Aradacanthia heissi (Hemiptera: Aradidae). Zootaxa. 3238(1):23–38.
- Song F, Li H, Shao R, Shi A, Bai X, Zheng X, Heiss E, Cai W. 2016. Rearrangement of mitochondrial tRNA genes in flat bugs (Hemiptera: Aradidae). Sci Rep. 6:25725.
- Sudhir K, Glen S, Koichiro T. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
