Abstract
Nasutitermes tiantongensis belongs to Nasutitermitinae and its mitochondrial genome was determined in this study. It consisted of 13 PCGs, 22 tRNAs, 2 rRNAs, and an A + T-rich control region, and its length was 15824 bp. The phylogenetic analysis suggested that the genus Nasutitermes was not monophyletic, and N. tiantongensis formed a sister group with Bultitermes laticephalus. The mitochondrial genome of N. tiantongensis provides a resource for evolutionary analysis within Nasutitermitinae.
Nasutitermitinae is a large subfamily of Termitidae, and of considerable economic importance (De Faria Santos et al. Citation2017). Nasutitermes belongs to the subfamily Nasutitermitinae, the genus status of many species within this subfamily is uncertain, and the species in the genus Nasutitermes is also complex and needed to be reclassification (Inward et al. Citation2007). Considering the complexity of the subfamily Nasutitermitinae, so we study their mitochondrial genomes of these species in this subfamily. Nasutitermes tiantongensis was first mentioned in 1993 (Zhou and Xu Citation1993). To date, there is no available information for the mitochondrial genome of N. tiantongensis. In this study, we firstly reported the complete genome sequence of this species.
Specimens were collected from Ningbo, China (29.86°N, 121.54°E) and deposited at College of Advanced Agricultural Sciences, Zhejiang A&F University (Contact person: Zhang Dayu, and email: [email protected]) under the voucher number NH-03. Total genomic DNA was extracted from the heads of 10 worker termites using FastPure Cell/Tissue DNA isolation Mini Kit (Vazyme, Nanjing, China) following the manufacturer’s instructions. Sequencing library was constructed after DNA extraction (Cloudna Technology, Beijing, China), and paired-end reads were sequenced using HiSeq XTen PE 150 of Illumina. The sequences were assembled and annotated using the MitoZ software (Meng et al. Citation2019). The total mitochondrial genome of N. tiantongensis was 15,824 bp in size, including 13 protein-coding genes (PCGs), 22 transfer RNA (tRNAs) genes, 2 ribosomal RNA (rRNAs) genes and a control region located between rrnS and trnI gene, and its gene arrangement was consistent with other termites (Lee et al. Citation2017). The minority strand (N-strand) encodes 14 genes (nad1, nad4, nad4L, nad5, trnQ, trnC, trnY, trnF, trnH, trnP, trnL1, trnV, rrnS and rrnL), and the other 23 genes are located at the majority strand (J-strand).
The total length of 13 PCGs of N. tiantongensis mitochondrial genome was 11,139 bp. All the PCGs were initiated with ATN, among which ATP8, ATP6, ND3 and ND6 used ATA as start codons, and the remaining used ATG as start codons. Except for nad1 with TAG, cox2 and nad5 with incomplete T as stop codon, the other genes were terminated with TAA. The mitochondrial genome of N. tiantongensis had 22 tRNAs with the typical clover-leaf secondary structure, aside from trnS1 lacking the dihydrouridine (DHU), which was common phenomenon observed in metazoan animals (Garey and Wolstenholme Citation1989). The rrnS was located between trnL and trnV, and rrnL was between trnV and control region. The control region was 1019 bp in size and had a high A + T content (71.93%).
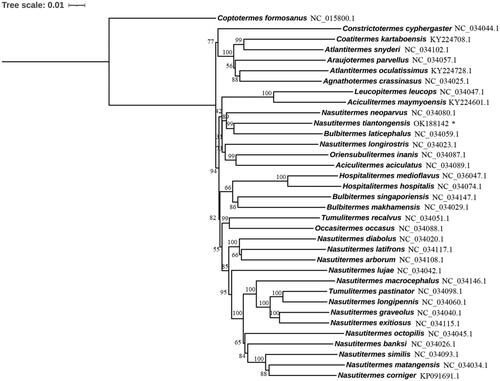
The phylogenetic tree based on nucleotide sequences of all PCGs from Nasutitermitinae species was constructed to investigate phylogenetic relationship. Maximum likelihood method was used to construct the phylogenetic tree using MEGA X, and the bootstrap value was set as 1000. The results suggested that the genus Nasutitermes was not monophyletic, and N. tiantongensis formed a sister group with Bultitermes laticephalus (). Previous phylogenetic analysis inferred from mitochondrial cytochrome oxidase II and 16S rRNA sequences also did not support the monophyly of the genus Nasutitermes (Bergamaschi et al. Citation2007). Phylogenetic relationships based on the analysis of three genes (cytochrome oxidase II, 12S rRNA and 28S rRNA) indicated that Nasutitermes was paraphyletic on the estimated cladogram and Nasutitermitinae were monophyletic (Inward et al. Citation2007). Hence, the genus Nasutitermes was paraphyletic. The species in this genus was complex, and worth for further study.
Ethical approval
Ethics approval was not required for this research.
Author contributions
Qian kexin and Gu Qijuan were involved in the conception and design, and the drafting of the paper; Qian Jiaojuan and Chen Haihong was involved in analysis and interpretation of the data; Zhang Dayu was involved in the conception and revising it critically for intellectual content; and all authors agree to the final approval of the version to be published and be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in Genbank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession number OK188142. The associated BioProject, SRA and Bio-Sample number are PRJNA835527, SRR19109539 and SAMN28104798, respectively.
Additional information
Funding
References
- Bergamaschi S, Dawes-Gromadzki TZ, Luchetti A, Marini M, Mantovani B. 2007. Molecular taxonomy and phylogenetic relationships among Australian Nasutitermes and Tumulitermes genera (Isoptera, Nasutitermitinae) inferred from mitochondrial COII and 16S sequences. Mol Phylogenet Evol. 45(3):813–821.
- De Faria Santos A, Fernandes Carrijo T, Marques Cancello E, Coletto Morales-Correa E. 2017. Phylogeography of Nasutitermes corniger (Isoptera: Termitidae) in the neotropical region. BMC Evol Biol. 17:230.
- Garey JR, Wolstenholme DR. 1989. Platyhelminth mitochondrial DNA evidence for early evolutionary origin of a tRNAse’AGN that contains a dihydrouridine arm replacement loop, and of serine-specifying AGA and AGG codons. J Mol Evol. 28(5):374–387.
- Inward D. J, Vogler AP, Eggleton P. 2007. A comprehensive phylogenetic analysis of termites (Isoptera) illuminates key aspects of their evolutionary biology. Mol Phylogenet Evol. 44(3):953–967.
- Lee W, Han T, Lee JH, Hong KJ, Park J. 2017. The complete mitochondrial genome of the subterranean termite, Reticulitermes speratus kyushuensis Morimoto, 1968 (Isoptera: rhinotermitidae). Mitochondrial DNA B Resour. 2(1):178–179.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Zhou B, Xu Y. 1993. A new species of Nasutitermes from Zhejiang Province, China (Isoptera: termitidae). Sci Technol Termites. 10(2):7.