Abstract
The Korean endemic earthworm Amynthas deogyusanensis Hong and James, 2001 (Clitellata: Megascolecidae) is found in the forest area of Deogyu Mountain, South Korea. In this study, the complete mitochondrial genome (mitogenome) of A. deogyusanensis was sequenced, assembled, and annotated. The mitogenome of A. deogyusanensis is a circular DNA molecule, consisting of 15,257 bp with an A + T content of 67.9%. It contains 13 protein-coding genes, two ribosomal RNA genes, 22 transfer RNA genes, and one non-coding region (control region). Phylogenetic analysis suggested that the family Megascolecidae is a monophyletic group with full support, whereas the genus Amynthas is non-monophyletic with the genera Metaphire and Duplodicodrilus.
Amynthas deogyusanensis Hong and James, Citation2001 is an earthworm endemic species to South Korea. The genus Amynthas Kinberg, 1867 is known to comprise more species than any other genus of the Pheretima complex (Sims and Easton Citation1972). Amynthas is the largest genus in family Megascolecidae and is one of the most abundant and diverse genus found in Korea and East Asia. Deogyu Mountain National Park located in Jeollabuk-do (central Korea), preserves the sub-alpine ecosystem. Amynthas deogyusanensis, belonging to the family Megascolecidae, is a species first discovered in the Deogyu Mountain, South Korea (Hong and James Citation2001). The body length, width, and segments of the species range from 102 to 110 mm, 5 to 5.7 mm, and 104 to 106 mm, respectively (Hong and James Citation2001). The male pore region of this species is unique and can be easily distinguished from the other endemic South Korean species ().
Figure 1. Clitellate of Amynthas deogyusanensis. (A) The dorsal view; (B) the clitellum annular XIV–XVI. The photograph from Deogyu Mountain, South Korea by Yong Hong on 2 August 2020.

The specimens of A. deogyusanensis were collected from Deogyu Mountain, Jeollabuk-do, South Korea (35°52′12.78″ N, 127°48′49.36″ E; 1296 m) on 2 August 2020. A specimen was deposited at Jeonbuk National University (Yong Hong, [email protected]) under the voucher number JBNU0004.
Total DNA was isolated from a single specimen using QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) and a sequencing library was constructed using Illumina TruSeq DNA Nano Library Prep Kit (Illumina Inc., San Diego, CA). The mitogenome sequence was generated by paired-end (2 × 150 bp) sequencing using Illumina HiSeq-X platform (San Diego, CA). The raw reads were assembled using SPAdes version 3.13.0 (Bankevich et al. Citation2012) based on the GenBank-registered reference mitogenome sequence of the earthworm A. pectiniferus (GenBank accession number: KT429018). Mitogenome annotation was conducted using MitoZ version 2.3 (Meng et al. Citation2019) and manually curated based on BLAST searches in the National Center for Biotechnology Information (NCBI) database. To explore the evolutionary relationships and phylogenetic position of A. deogyusanensis, the available mitogenome sequences of the 27 species in the Megascolecidae family were collected from the NCBI database (Boore and Brown Citation1995; Wang et al. Citation2015; Zhang et al. Citation2016a, Citation2016b, Citation2019; Hong et al. Citation2017). Mitogenome sequences from the Lumbricidae and Moniligastridae families were used as outgroups. Nucleotide sequences of 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), and two ribosomal RNA genes (rRNAs) from each mitogenome were aligned using the clustal omega tool in Geneious Primer 2021, and the aligned sequences were concatenated into a dataset. The phylogenetic tree was constructed by the Bayesian inference (BI) method using MrBayes v3.2.7 (Ronquist et al. Citation2012). Markov chain Monte Carlo analysis was performed for 1,000,000 generations (the average standard deviation of split frequencies was 0.007). The first 25% of the tree corresponding to the ‘burn-in’ period was discarded and the remaining parts of the tree were used to construct the majority-rule consensus tree.
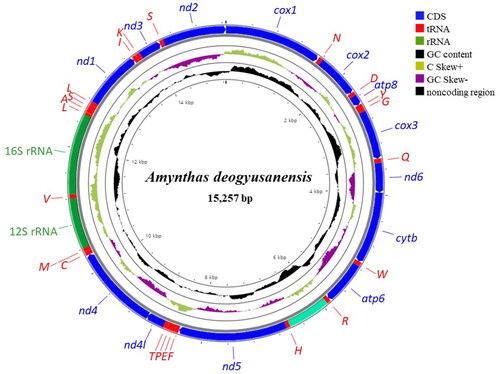
The complete mitogenome of A. deogyusanensis is a circular DNA molecule consisting of 15,257 bp with 13 PCGs, 22 tRNAs, two rRNAs, and one major non-coding control region (). The arrangement of mitochondrial genes of A. deogyusanensis is identical to that of other available mitogenomes of megascolecids species (Boore and Brown Citation1995; Wang et al. Citation2015; Zhang et al. Citation2016a, Citation2016b, Citation2019). We observed that all 13 PCGs start with an ATG codon, which is typical for invertebrate mitochondrial PCGs. Seven PCGs end with an complete stop codons, TAA and TAG, and six PCGs end with an incomplete stop codon, T. The A + T content of the whole mitogenome is 67.9%, which is similar to that found in the megascolecid species (61.6–67.2%). A 679-bp fragment of the putative control region is located at the junction between trnR and trnH.
Figure 2. Circular sketch map of the Amynthas deogyusanensis mitogenome. The mitogenome map was generated with CGView server (http://cgview.ca). Each transfer RNA gene is represented by a one-letter amino acid code. Different colors represent different gene blocks.
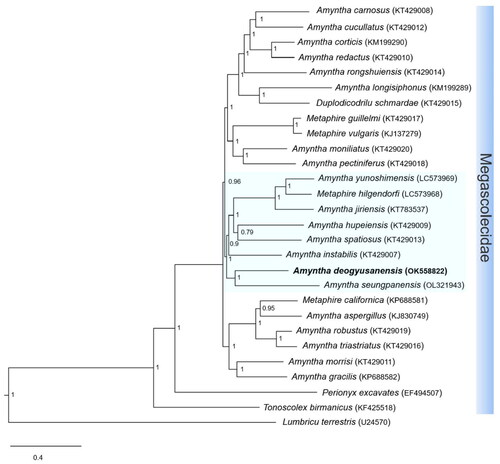
Phylogenetic analysis suggested that A. deogyusanensis is a sister group of A. yunoshimensis, M. hilgendorfi, A. jiriensis, A. spatiosus, A. hupeiensis, A. instabilis, and A. seungpanensis in the family Megascolecidae (). This relationship presents high nodal support in BI analysis. Additionally, the family Megascolecidae presents a monophyletic group with the highest nodal support, whereas Amynthas appears as a non-monophyletic group with the genera Metaphire and Duplodicodrilus (Hong et al. Citation2017; Zhang et al. Citation2019).
Figure 3. Phylogenetic tree of the 27 species of the Megascolecidae family. Phylogenetic analysis was done using Bayesian inference (BI) method, based on whole mitogenome sequences, excluding the non-coding region. The numbers at each node specify Bayesian posterior probabilities (BPP) by BI. The scale bar indicates the number of substitutions per site. Lumbricus terrestris (Boore and Brown Citation1995) is used as the outgroup. GenBank accession numbers are mentioned next to species names.
Ethical approval
The samples used in this study are earthworms that are not included in the list of protected animals, and hence, the ethical statement is not applicable.
Author contributions
Conceptualization, Y.H.; methodology, J.C.K. and Y.H.; data analysis, J.C.K.; investigation, J.C.K.; resources, Y.H.; data curation, J.C.K. and Y.H.; original draft preparation, Y.H. and J.C.K.; review and editing, Y.H.; project administration, Y.H.; funding acquisition, Y.H. All authors have read and agreed to the published version of the manuscript.
Disclosure statement
The authors report there are no competing interests to declare.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/OK558822, under the accession no. OK558822. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA769829, SRR16507334, and SAMN22374388, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Boore JL, Brown WM. 1995. Complete sequence of the mitochondrial DNA of the annelid worm Lumbricus terrestris. Genetics. 141(1):305–319.
- Hong Y, James SW. 2001. New species of Korean Amynthas Kinberg, 1867 (Oligochaeta, Megascolecidae) with two pairs of spermathecae. Rev Suisse Zool. 108:65–93.
- Hong Y, Kim MJ, Wang AR, Kim IK. 2017. Complete mitochondrial genome of the earthworm, Amynthas jiriensis (Clitellata: Megascolecidae). Mitochondrial DNA A DNA Mapp Seq Anal. 28(2):163–164.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Sims RW, Easton EG. 1972. A numerical revision of the earthworm genus Pheretima auct. (Megascolecidae: Oligochaeta) with the recognition of new genera and an appendix on the earthworms collected by the Royal Society North Borneo Expedition. Biol J Linnean Soc. 4:169–268.
- Wang AR, Hong Y, Win TM, Kim I. 2015. Complete mitochondrial genome of the Burmese giant earthworm, Tonoscolex birmanicus (Clitellata: Megascolecidae). Mitochondrial DNA. 26:467–468.
- Zhang L, Jiang J, Dong Y, Qiu J. 2016a. Complete mitochondrial genome of a pheretimoid earthworm Metaphire vulgaris (Oligochaeta: Megascolecidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):297–298.
- Zhang L, Jiang J, Dong Y, Qiu J. 2016b. Complete mitochondrial genome of an Amynthas earthworm, Amynthas aspergillus (Oligochaeta: Megascolecidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:1876–1877.
- Zhang Q, Liu H, Zhang Y, Ruan H. 2019. The complete mitochondrial genome of Lumbricus rubellus (Oligochaeta, Lumbricidae) and its phylogenetic analysis. Mitochondrial DNA B Resour. 4(2):2677–2678.
