Abstract
Holarrhena pubescens Wall. ex G. Don, 1837 is an important medicinal plant belonging to the Holarrhena genus in the Apocynaceae family. In this study, the complete chloroplast (cp) genome sequence of H. pubescens was sequenced using the Illumina NovaSeq platform. The cp genome of H. pubescens was 160,108 bp in length with 37.21% overall GC content. The cp genome of H. pubescens containing a large single-copy region (LSC, 88,685 bp), a small single-copy region (SSC, 18,671 bp), and a pair of inverted repeat regions (SSC, 26,376 bp). The cp genome encoded 129 genes, including 84 protein-coding genes, 37 tRNA genes, and eight rRNA genes. Phylogenetic analysis based on complete protein coding genes sequences revealed that H. pubescens was closest to Beaumontia murtonii.
Introduction
Holarrhena pubescens Wall. ex G.Don, 1837 is an important medicinal plant belonging to the Holarrhena genus in the Apocynaceae family (Khan et al. Citation2021; Namasudra et al. Citation2021), native to central and southern Africa, the Indian subcontinent, Indochina, and parts of China (Gupta et al. Citation2021). The bark of H. pubescens has been widely used to treat malaria, cough, cold, bronchopneumonia, dyspepsia, and diarrhea (Sharma et al. Citation2015; Gupta et al. Citation2021; Khan et al. Citation2021; Namasudra et al. Citation2021). H. pubescens is considered the most valuable medicine due to its numerous medicinal properties and nontoxic side effects (Zahara et al. Citation2020; Bala et al. Citation2022). Previous studies have focused on its pharmacological activity and chemical composition (Khan et al. Citation2021; Sripahco et al. Citation2021; Chowdhury and Mazumdar Citation2022). However, there is no report on the genomic studies of H. pubescens. Therefore, we present the complete chloroplast (cp) genome of H. pubescens for the first time, which will provide a theoretical and data foundation for the effective development, protection, and utilization of the H. pubescens.
Materials and methods
Plant material and DNA sequencing
This article is licensed under the Yunnan Province Biodiversity Conservation Regulations and with permission from Xishuangbanna (Yunnan Province, China), and the Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences (Beijing, China). Fresh leaves of H. pubescens were collected from Xishuangbanna City (N21°55′40″, E101°15′12″), Yunnan Province, China (). We have obtained permission from the local forestry department to collect samples. This study was conducted in accordance with the laws of the People’s Republic of China. This specimen was identified by Kunming Cai Zhi Biotechnology Co. (Kunming, China). The voucher specimen was deposited in the herbarium of the Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences (Xiangxiao Meng, [email protected]) under the voucher number: XSBN20220524. Genomic DNA was extracted using the Plant Genomic DNA Kit (Tiangen, Beijing, China). After the sample is tested as qualified, take 0.5 μg for each sample DNA template, according to the manufacturer’s instructions (Illumina NovaSeq 6000), construct a short fragment insertion library (insertion size 350–500 bp) for sequencing, run the paired-end sequencing program (PE), and obtain 150 bp paired-end sequencing reads.
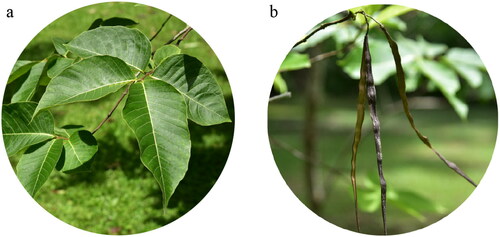
Figure 1. (a) The leaves of H. pubescens. (b) The follicles of H. pubescens. The leaf blade of this species is ovate or elliptic, with 10–15 pairs of lateral veins, and the follicles of this species are linear, with white, punctate lenticels. The photos of H. pubescens were taken by the authors in Xishuangbanna county, Yunnan Province, China.
Genome assembly and annotation
The high-quality reads were assembled with GetOrganelle v.1.7.5 (Jin et al. Citation2020) and annotated by CPGAVAS2 (Shi et al. Citation2019). The annotated cp genome was deposited in the Genome Warehouse (accession number: GWHBKKE01000000).
Phylogenetic analysis
To evaluate the phylogenetic position of H. pubescens, 11 complete cp genome sequences from Apocynaceae and one outgroup species (Gentiana crassicaulis) were downloaded from the NCBI database. The complete protein coding genes of 13 species were extracted and aligned by MAFFT v7.471 (Katoh and Standley Citation2013). Maximum-likelihood (ML) analyses were performed with RAxML v8.2.12 (Stamatakis Citation2014) using the rapid bootstrap algorithm with 1000 replicates to assess branch support, combined with a search of the best-scoring ML tree under default parameters.
Results and discussion
Genome structure analysis
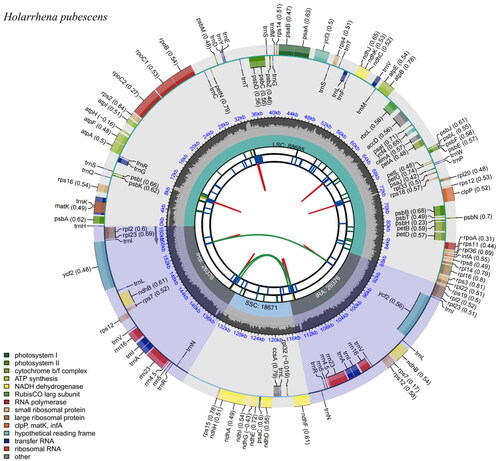
In total, 10.4 Gb of raw data were obtained (69,706,856 reads). The sequences obtained from sequencing are filtered with low quality data to obtain clean reads (67,006,166 reads), and based on the clean reads, an appropriate amount of data is used for assembly and subsequent analysis to finally obtain the complete genome sequence, the genome assembly coverage is 100% (Figure S1). The complete cp genome of H. pubescens is 160,108 bp in length () and includes a pair of inverted repeats 26,376 bp long, separated by a small and a large single-copy region of 18,671 bp and 88,685 bp, respectively. A total of 129 genes with 84 coding sequences, 37 tRNA genes, and eight unique rRNA sequences were identified. Among them, the ndhD gene is an RNA editing gene, using ACG as the start codon, which was predicted based on the start codon and the length of the gene of the ndhD gene of the closely related species Nerium oleander L. (NC_025656). The GC content of the cp genome was 37.21%.
Figure 2. Chloroplast reference genome of Holarrhena pubescens. This figure has six circles from the center to the outside. The first circle shows the forward and reverse repeats connected with the red and green arcs, respectively. The second circle and the third circle show tandem repeats and microsatellite sequences marked with short strips, respectively. The fourth circle indicates the position of LSC, SSC, IRA, and IRB, and the fifth circle indicates the GC content. The genes outside the sixth circle are transcribed counterclockwise, while the genes inside are transcribed clockwise. Genes belonging to different functional groups are color coded, as shown in the left corner.
Phylogenetic analysis
Phylogenetic analysis revealed that H. pubescens was closely related to Beaumontia murtonii Craib (). The cp genome sequence of H. pubescens in this study might provide important information for phylogenetic and evolutionary studies in Apocynaceae.
Figure 3. Phylogenetic analysis of 12 species and one taxon as outgroups based on complete protein coding genes sequences by RAxML, bootstrap support value near the branch. The following sequences were used: MK783315 Trachelospermum jasminoides (Wang et al. Citation2019), NC_056322 Chonemorpha megacalyx (Deng et al. Citation2021), MG963251 Beaumontia murtonii (Fishbein et al. Citation2018), GWHBKKE01000000 Holarrhena pubescens, NC_025656 Nerium oleander (Straub et al. Citation2014), NC_054286 Wrightia laevis (Li et al. Citation2020), NC_033354 Carissa macrocarpa (Jo et al. Citation2017), NC_051546 Cerbera manghas (Liao et al. Citation2020), NC_047244 Rauvolfia serpentina (Zhang et al. Citation2021), NC_046841 Rauvolfia verticillata (Chen et al. Citation2019), NC_021423 Catharanthus roseus (Ku et al. Citation2013), NC_065203 Vinca major (Hendrian Citation2007), and NC_027442 Gentiana crassicaulis (Ni et al. Citation2017).

Author contributions
Rushuang Xiang analyzed the data, prepared figures, authored drafts of the paper, and approved the final draft. Sijia Wang analyzed the data. Huihua Wan designed the experiments, contributed reagents/materials/analysis tools, revised the content, and approved the final draft. All authors agree to be accountable for all aspects of the work.
Supplemental Material
Download MS Word (1,012.5 KB)Disclosure statement
The authors reported no potential conflict of interest.
Data availability statement
The genome sequence data that support the findings of this study are openly available in the Genome Warehouse of NGDC at https://ngdc.cncb.ac.cn/gwh under the accession number: GWHBKKE01000000. The associated BioProject, GSA, and Bio-Sample numbers are PRJCA011728, CRA008103, and SAMC873599, respectively.
Additional information
Funding
References
- Bala K, Melkani I, Singh AP, Singh AP, Kaur J. 2022. Holarrhena antidysenterica in inflammatory bowel disease: a potential review. J Drug Deliv Ther. 12(4):221–226.
- Chen W, Liang W, Li A, Ma J. 2019. Characterization of the complete plastid genome of Rauvolfia verticillata (Apocynaceae), with its phylogenetic analysis. Mitochondrial DNA B Resour. 4(2):4190–4191.
- Chowdhury SK, Mazumdar T. 2022. The pesticidal activities of Rhizospheric Bacteria isolated from Holarrhena pubescens, their plant growth promotion and IAA production optimization. Sci J Biol. 1(5):10–21.
- Deng YP, Zhang XL, Li LY, Yang T, Liu GH, Fu YT. 2021. Characterization of the complete mitochondrial genome of the swine kidney worm Stephanurus dentatus (Nematoda: Syngamidae) and phylogenetic implications. Vet Parasitol. 295:109475.
- Fishbein M, Livshultz T, Straub S, Simoes AO, Boutte J, McDonnell A, Foote A. 2018. Evolution on the backbone: Apocynaceae phylogenomics and new perspectives on growth forms, flowers, and fruits. Am J Bot. 105(3):495–513.
- Gupta N, Choudhary SK, Bhagat N, Karthikeyan M, Chaturvedi A. 2021. In silico prediction, molecular docking and dynamics studies of steroidal alkaloids of Holarrhena pubescens Wall. ex G. Don to guanylyl cyclase C: implications in designing of novel antidiarrheal therapeutic strategies. Molecules. 26(14):4147.
- Hendrian KK. 2007. Molecular phylogeny of Ochrosia sensu lato (Apocynaceae) based on rps16 intron and ITS sequence data: supporting the inclusion of Neisosperma. Chromosome Bot. 4(2):133–140.
- Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Jo S, Kim HW, Kim YK, Cheon SH, Kim KJ. 2017. The complete plastome sequence of Carissa macrocarpa (Eckl.) A. DC. (Apocynaceae). Mitochondrial DNA B Resour. 2(1):26–28.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Khan S, Viquar U, Alam MA, Moin MS, Khatoon F, Minhajuddin A. 2021. Ethno-pharmacology of Holarrhena antidysenterica wall. ex g. don. (Tewāj) in light of unani system of medicine. Int J Bot Stud. 5(6):1133–1139.
- Ku C, Chung WC, Chen LL, Kuo CH. 2013. The complete plastid genome sequence of madagascar periwinkle Catharanthus roseus (L.) G. Don: plastid genome evolution, molecular marker identification, and phylogenetic implications in asterids. PLoS One. 8(6):e68518.
- Li LM, Fu JX, Song XQ. 2020. Complete plastome sequence of Wrightia laevis Hook. f. a dyestuff species. Mitochondrial DNA B Resour. 3(5):2533–2534.
- Liao M, Wei XF, Ding HP, Tang GD. 2020. The complete chloroplast genome of the highly poisonous plant Cerbera manghas L. (Apocynaceae). Mitochondrial DNA B Resour. 5(3):3084–3085.
- Namasudra S, Phukan P, Bawari M. 2021. GC–MS analysis of bioactive compounds and safety assessment of the ethanol extract of the barks of Holarrhena pubescens Wall. ex.G.Don (family Apocynaceae): sub-acute toxicity studies in swiss albino mice. Pharmacogn J. 13(1):162–171.
- Ni L, Zhao Z, Xu H, Chen S, Dorje G. 2017. Chloroplast genome structures in Gentiana (Gentianaceae), based on three medicinal alpine plants used in Tibetan herbal medicine. Curr Genet. 63(2):241–252.
- Sharma DK, Gupta VK, Kumar S, Joshi V, Mandal RS, Prakash AG, Singh M. 2015. Evaluation of antidiarrheal activity of ethanolic extract of Holarrhena antidysenterica seeds in rats. Vet World. 8(12):1392–1395.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Sripahco T, Tovaranonte J, Pripdeevech P. 2021. Chemical composition and antimicrobial activity of essential oil of Holarrhena pubescens flowers. Chem Nat Compd. 57(4):781–783.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Straub SC, Moore MJ, Soltis PS, Soltis DE, Liston A, Livshultz T. 2014. Phylogenetic signal detection from an ancient rapid radiation: effects of noise reduction, long-branch attraction, and model selection in crown clade Apocynaceae. Mol Phylogenet Evol. 80:169–185.
- Wang H, Cheng X, Chen W, Li L, Chen L. 2019. Complete plastome sequence of Trachelospermum jasminoides (Lindley) Lemaire (Apocynaceae). Mitochondrial DNA B Resour. 1(4):2086–2087.
- Zahara K, Panda SK, Swain SS, Luyten W. 2020. Metabolic diversity and therapeutic potential of Holarrhena pubescens: an important ethnomedicinal plant. Biomolecules. 10(9):1341.
- Zhang K, Liu L, Shan X. 2021. Characterization of the complete chloroplast genome of Tabernaemontana divaricata (Apocynaceae), a valuable and endangered plant. Mitochondrial DNA B Resour. 6(11):3125–3126.
