Abstract
The Siberian Rubythroat (Calliope calliope), a shy and cautious passerine bird, is widely distributed throughout Siberia, Asia, and Europe. In this study, the complete mitogenome of the Siberian Rubythroat samples collected in Heilongjiang, China, was sequenced. The whole length of the complete mitochondrial genome is 16,840 bp, consisting of 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and one control region. Only one overlapping gene was found among the 13 protein-coding genes (ND4L/ND4). The length of the control region is 1068 bp. The nucleotide composition is 29.32% A, 23.05% T, 14.80% G, and 32.82% C. Phylogenetic analysis indicated a close genetic relationship between C. calliope and Luscinia cynaura.
1. Introduction
1.1. Background
The Siberian Rubythroat (Calliope calliope Pallas 1776) is a small insectivorous bird that is widely distributed throughout Siberia and the Russian Far East (Ivanitskii and Monakhova Citation2020). Because of its melodious song, C. calliope is a valuable cage bird in China. The genome of C. calliope has not been published so far.
1.2. Justification
C. calliope is listed as Near Threatened according to the Red List of Threatened Species. The destruction of habitats preferred by these birds across their range is assumed to be the main cause of its uneven distribution and population decline. Moreover, the introduction of non-native species and climate change may threaten its survival. To better conserve this species, the complete mitogenome of C. calliope was sequenced.
2. Materials
The complete mitochondrial genome of C. calliope was sequenced using muscle tissue collected from samples in Heilongjiang, China. The specimens were deposited with the College of Wildlife and Protected Area, Northeast Forestry University, Harbin, China (No. BYQY201004; Zhensheng Liu: [email protected]). C. calliope, a member of the Muscicapidae family, selects forested habitats dominated by Erman’s birch and dwarf Siberian pine. They prefer shrub areas of low height that provides good cover (Xin et al. 2002).
3. Methods
Total genomic DNA was extracted and sequenced using a REPLI-g Mitochondrial DNA Kit on the Illumina Hiseq sequencing platform by Nanjing Personal Gene Technology Co. Ltd., China (Ru et al. Citation2021). High quality reads were then de novo assembled using MitoZ (Meng et al. Citation2019). The mitotic genome was annotated using the MITOS web server (Bernt et al. Citation2013). The phylogenetic relationship was inferred using the Neighbor-Joining method (Saitou and Nei Citation1987), performed in MEGA11 (Kumar et al. Citation2016). The bootstrap consensus tree, inferred from 1,000 replicates, was used to represent the evolutionary history of analyzed taxa (Felsenstein Citation1985).
4. Results and discussion
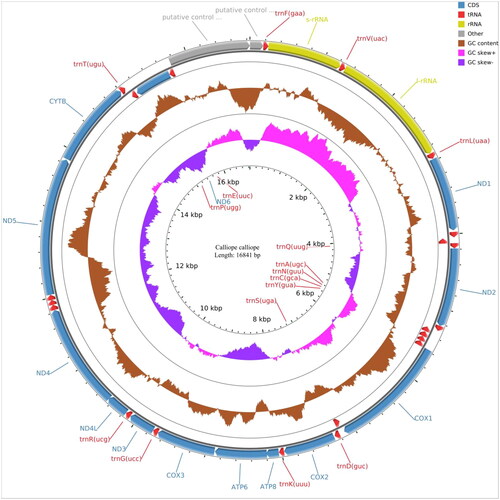
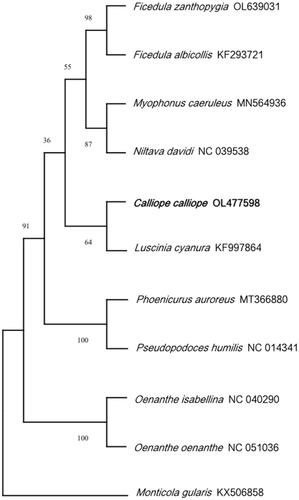
The mitochondrial genome is composed of 13 protein-coding genes, 2 rRNA genes (s-rRNA and l-rRNA), 22 tRNA genes, and 1 control region. The whole genome has a length of 16,840 bp, with a base composition of 29.32% A, 23.05% T, 14.80% G, and 32.82% C. The 13 protein-coding genes have a length of 10,997 bp, and all genes are encoded on the same strand, except for ND6, which is encoded on the light strand. All seven protein-coding genes start with ATG (COX3, ND1, ND3, ND4, ND4L, ATP6, and ATP8) except for ND2, ND5, and Cytb (ATC start codon), ND6 (TAA start codon), and both COX1 and COX2 (GTG start codon). The total length of all tRNA genes is 1544 bp, ranging from 66 bp (tRNA-Ser and tRNA-Cys) to 76 bp (tRNA-Trp). The lengths of the two rRNA genes and the control region are 985 bp (s-rRNA), 1599 bp (l-rRNA), and 1068 bp (control region). The phylogenetic tree indicates that C. calliope is most closely related to Luscinia cynaura.
In conclusion, the complete mitochondrial genome of C. calliope provides valuable information for further genetic studies, thus contributing to improvements of conservation and management of C. calliope ().
Ethical approval
The muscle tissue of the Siberian Rubythroat was extracted from the individual which died naturally in the wild not more than three days old. Ethical approval is not required in this case.
Authors’ contributions
Conception and design, Zhensheng Liu, Liwei Teng, and Xiaoyu Zhou; Specimen collection and identification, Dehuai Meng; Data analysis, Yao Zhao and Nan Zhang; Writing-Original Draft Preparation, Yao Zhao and Nan Zhang; Writing-Review, Nan Zhang. All authors agree to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/, reference number OL477598. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA781062, SRR16962849, and SAMN23227665, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Felsenstein J. 1985. Phylogenies and the comparative method. Am Nat. 125(1):1–15.
- Ivanitskii V, Monakhova M. 2020. The song structure of the Siberian Rubythroat Luscinia Calliope calliope: branching syntax underlies complex and variable vocalization. Ornithological Science. 19(2):177–186.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Rhim S-J, Hur W-H, Lee C-B, Park Y-S, Choi S-Y, Lee W-S. 2002. Characteristics of vegetation structure in breeding area of Siberian rubythroat (Luscinia calliope) in Daecheongbong peak, Mt. Seoraksan national park, South Korea. J for Res. 13(3):239–240.
- Ru YY, Yi L, Zhao MZ. 2021. The complete mitochondrial genome of Tetramorium tsushimae (Emery, 1925) (Hymenoptera: formicidae). Mitochondeial DNA Part B. 7(1):40–42.
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425.