Abstract
In this study, we sequenced and analyzed the complete mitogenome of Smerinthus caecus Ménétriés, 1857. The mitogenome of S. caecus is a circular structure, and 15,363 bp long in size and encodes 13 protein-coding genes (PCGs), two ribosomal RNA genes (rRNAs), 22 transfer RNA genes (tRNAs), and a control region (CR). An extremely high AT bias of 79.2% was found in the nucleotide composition of mitogenome. Most of the PCGs used ATN as the start codon and TAA or TAG as the stop codon, which is similar to most other insect mitogenomes, except cox1, which starts with CGA. The phylogeny of Smerinthinae was reconstructed using a maximum-likelihood method, a total of 33 mitogenomes were sampled for phylogenetic analyses. The subfamily Langiinae was selected as outgroup. The results confirmed the position of S. caecus in the Smerinthinae, in which Smerinthus caecus was placed as the sister taxon to Smerinthus planus, then to Laothoe amurensis.
The Lepidoptera are the second largest order of Insecta after Coleoptera, with more than 157,000 described species in 43 superfamilies (Mitter et al. Citation2017). The hawkmoths (family Sphingidae) belong to superfamily Bombycoidea and currently includes more than 1600 species (Kitching et al. Citation2018). However, our current knowledge of hawkmoth mitogenomes is still limited. Moreover, the deep-level lepidopteran phylogeny remains poorly understood. The hawkmoth that is the subject of the current study, Smerinthus caecus Ménétriés, 1857 is distributed in northeast Asia and recently spread to Europe from Siberia (Pittaway and Kitching Citation2022). In this study, we sequenced the hitherto unknown complete mitochondrial genome of S. caecus to reconstruct a phylogenetic tree of Smerinthinae (Lepidoptera: Sphingidae).
A male specimen of S. caecus was collected from Dabie Mountain, Lu’an City, Anhui Province, China (31°13′08″N, 116°20′19″E) in May 2021 and deposited in the Entomological Museum, College of Life Sciences, Anhui Normal University (https://www.ahnu.edu.cn/, YX, Huang, [email protected]) under the accession no. DB20210522. It was identified on the basis of wing pattern and the morphology of its genitalia. A whole genome shotgun (WGS) strategy was used with sequencing undertaken on the Illumina platform. The raw paired reads were quality-trimmed and assembled into the complete circular mitogenome in Novoplasty 2.7.2 (Nicolas et al. Citation2017).
The complete mitogenome of S. caecus (GenBank accession number: MZ593603) is a circular structure, and 15,363 bp long. The typical 37 mitochondrial genes (13 PCGs, 22 transfer RNA genes (tRNAs), and two ribosomal RNA genes (rRNAs)) and an A + T-rich region were present. The overall base composition of the mitogenome was calculated to be A: 40.4%, T: 38.8%, C: 12.8%, and G: 8.0%. The nucleotide composition had an obvious adenine and thymine bias, and the A + T content was 79.2%. Of the 37 genes, 23 (nine PCGs and 14 tRNAs) were encoded on the majority strand (J-strand), and the remaining 14 were located on minority strand (N-strand). Nine of the 13 PCGs were encoded on the J-strand (nad2, nad3, nad6, cox1, cox2, cox3, atp6, atp8, cytb), and the other four were located on the N-strand. The total length of the 13 PCGs in the S. caecus mitogenome was 12,504 bp, accounting for approximately 81.4% of the whole mitogenome. Most of the protein-coding genes (PCGs) used ATN as the start codon and TAA or TAG as the stop codon, which is similar to most other insect mitogenomes, except cox1, which starts with CGA (Crozier and Crozier Citation1993; Korkmaz et al. Citation2015). Having cox1 genes that start with CGA is common among Lepidopterans, especially for all the species in Sphingidae (Wang et al. Citation2021; Chen et al. Citation2022). The S. caecus mitogenome contained the typical 22 tRNAs with lengths ranging from 64 bp (trnC) to 71 bp (trnK). Among them, eight tRNAs (trnQ, trnC, trnY, trnF, trnH, trnP, trnL, trnV) were encoded by the N-strand and the remaining 14 were encoded by the J-strand. The two rRNA genes were identified as being on the N-strand in the S. caecus mitogenome.
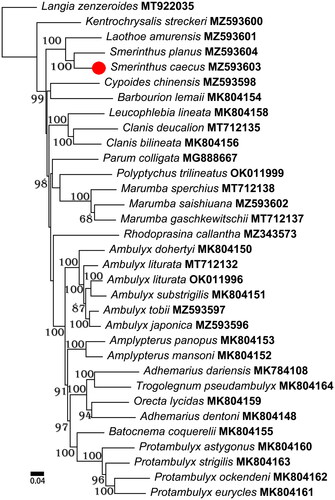
To investigate the phylogenetic implications of the S. caecus mitogenome in Smerinthinae phylogeny, a total of 33 mitogenomes were sampled for phylogenetic analyses. The subfamily Langiinae was selected as outgroup. Nucleotide sequences from each PCG were aligned by MUSCLE nested within MEGA X (Sudhir et al. Citation2018). Alignments of individual genes were then concatenated as a combined matrix with DAMBE 5.3.74 (Xia Citation2013). We analyzed the nucleotide sequences of the PCGs using maximum-likelihood (ML) implemented on the W-IQ-Tree web server to reconstruct the phylogenetic relationships of S. caecus with regard to other sphingids under the best fit substitution models (Chernomor et al. Citation2016; Trifinopoulos et al. Citation2016; Kalyaanamoorthy et al. Citation2017; Minh et al. Citation2020). An ultrafast bootstrap (UFB) of 1000 replications was used in this analysis to assess branch supports (Hoang et al. Citation2018). The results () confirmed the position of S. caecus in the Smerinthinae, in which Smerinthus caecus was placed as the sister taxon to Smerinthus planus, then to Laothoe amurensis.
Figure 1. Phylogenetic relationships within Smerinthinae based on the sequences of 13 protein-coding genes analyzed using maximum-likelihood methods. The values of an ultrafast bootstrap (UFB) of 1000 replications are shown on the nodes. The following records were used: ‘Adhemarius dariensis MK784108, Adhemarius dentoni MK804148, Ambulyx dohertyi MK804150, Ambulyx substrigilis MK804151, Amplypterus mansoni MK804152, Amplypterus panopus MK804153, Barbourion lemaii MK804154, Batocnema coquerelii MK804155, Clanis bilineata MK804156, Leucophlebia lineata MK804158, Orecta lycidas MK804159, Protambulyx astygonus MK804160, Protambulyx eurycles MK804161, Protambulyx ockendeni MK804162, Protambulyx strigilis MK804163, and Trogolegnum pseudambulyx MK804164 (Timmermans et al. Citation2019); Ambulyx liturata MT712132, Clanis deucalion MT712135, Langia zenzeroides MT922035, Marumba gaschkewitschii MT712137, and Marumba sperchius MT712138 (Wang et al. Citation2021); Kentrochrysalis streckeri MZ593600 (Huang et al. Citation2022); Smerinthus planus MZ593604 (Meng et al. Citation2022a); Ambulyx tobii MZ593597 (Meng et al. Citation2022b); Laothoe amurensis MZ593601 (Sun et al. Citation2022); Smerinthus caecus MZ593603 (this study); Ambulyx japonica MZ593596, Ambulyx liturata OK011996, Cypoides chinensis MZ593598, Marumba saishiuana MZ593602, Parum colligata MG888667, Polyptychus trilineatus OK011999, and Rhodoprasina callantha MZ343573 (Unpublished)’.
Author contributions
Yin-Feng Meng: study concept and design, analysis and interpretation of data, drafting and critical revision of the manuscript. Chao-Fan Chen: study concept and design, analysis and interpretation of data. Yi-Xin Huang: study concept and design, analysis and interpretation of data. Xu Wang: study concept and design, analysis and interpretation of data. Bo Zhang: study concept and design, analysis and interpretation of data, drafting and critical revision of the manuscript, and final approval of the version to be published. All authors agree to be accountable for all aspects of the work.
Ethics statement
No specific permits were required for the insect specimens collected for this study. The field studies did not involve endangered or protected species. The insect species sequenced is a common hawkmoth species in China and is not included in the 'List of Protected Animals in China’.
Acknowledgements
We thank Ian J. Kitching and Ping Shin Lee to review the manuscript.
Disclosure statement
The authors declare no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The data supporting this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/genbank/, reference number MZ593603. The associated BioProject, Bio-Sample numbers, and SRA are PRJNA752517, SAMN20600166, and SRR15358143, respectively.
Additional information
Funding
References
- Chen Q, Chen L, Liao CQ, Wang X, Wang M, Huang GH. 2022. Comparative mitochondrial genome analysis and phylogenetic relationship among lepidopteran species. Gene. 830:146516.
- Chernomor O, von Haeseler A, Minh BQ. 2016. Terrace aware data structure for phylogenomic inference from supermatrices. Syst Biol. 65(6):997–1008.
- Crozier RH, Crozier YC. 1993. The mitochondrial genome of the honeybee Apis mellifera: complete sequence and genome organization. Genetics. 133(1):97–117.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522.
- Huang YX, Zhu XS, Zhang H, Qi LQ, Jin HZ, Bian CL, Chen WL, Wang X. 2022. Complete mitochondrial genome of Kentrochrysalis streckeri (Lepidoptera: Sphingidae) and phylogenetic analysis. Mitochondrial DNA B Resour. 7(6):908–910.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Kitching IJ, Rougerie R, Zwick A, Hamilton CA, St Laurent RA, Naumann S, Ballesteros Mejia L, Kawahara A. 2018. A global checklist of the Bombycoidea (Insecta: Lepidoptera). Biodivers Data J. 6:e22236.
- Korkmaz EM, Doğan Ö, Budak M, Başıbüyük HH. 2015. Two nearly complete mitogenomes of wheat stem borers, Cephus pygmeus (L.) and Cephus sareptanus Dovnar-Zapolskij (Hymenoptera: Cephidae): an unusual elongation of rrnS gene. Gene. 558(2):254–264.
- Meng YF, Chen CF, Huang YX, Wang X, Zhang B. 2022a. Phylogenetic relationship and characterization of the complete mitochondrial genome sequence of Smerinthus planus (Lepidoptera: Sphingidae). Mitochondrial DNA B Resour. 7(6):941–943.
- Meng YF, Lv GT, Huang YX, Wang X, Wu YL. 2022b. The complete mitochondrial genome sequence of the hawkmoth, Ambulyx tobii (Lepidoptera: Sphingidae) and phylogenetic analysis. Mitochondrial DNA B Resour. 7(4):629–631.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Mitter C, Davis DR, Cummings MP. 2017. Phylogeny and evolution of Lepidoptera. Annu Rev Entomol. 62:265–283.
- Nicolas D, Patrick M, Guillaume S. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Pittaway AR, Kitching IJ. 2022. Sphingidae of the Eastern Palaearctic (including Siberia, the Russian Far East, Mongolia, China, Taiwan, the Korean Peninsula and Japan); [accessed 2022 May 21]. https://tpittaway.tripod.com/china/china.htm.
- Sudhir K, Glen S, Li M, Christina K, Koichiro T. 2018. Mega X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):6.
- Sun Y, Wang J, Wang X. 2022. Sequencing and analysis of the complete mitochondrial genome of Laothoe amurensis sinica (Lepidoptera: Sphingidae) from China and its phylogenetic analysis. Mitochondrial DNA B Resour. 7(5):750–752.
- Timmermans MJ, Daghmoumi SM, Glass D, Hamilton CA, Kawahara AY, Kitching IJ. 2019. Phylogeny of the Hawkmoth Tribe Ambulycini (Lepidoptera: Sphingidae): mitogenomes from museum specimens resolve major relationships. Insect Syst Divers. 3(6):12.
- Trifinopoulos J, Nguyen LT, Haeseler AV, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235.
- Wang X, Zhang H, Kitching I, Xu ZB, Huang YX. 2021. First mitogenome of subfamily Langiinae (Lepidoptera: Sphingidae) with its phylogenetic implications. Gene. 789:145667.
- Xia X. 2013. DAMBE5: a comprehensive software package for data analysis in molecular biology and evolution. Mol Biol Evol. 30(7):1720–1728.
