Abstract
The differentiation between Neolissochilus and Acrossocheilus species based only on morphology is ambiguous; however, phylogenetic analysis using their mitogenome sequences provides conclusive results. Here, the phylogenetic position of Neolissochilus hendersoni (Herre, 1940) was determined using its mitogenome data. Total DNA from N. hendersoni was sequenced using the Illumina NovaSeq6000 platform, and annotation of mitochondrial genes was performed using MITOS2. Phylogenetic trees were constructed using the complete mitogenomes of 16 fish species. The mitogenome of N. hendersoni was found to be 16584 bp long, containing two ribosomal RNA genes, 13 protein-coding genes, 22 transfer RNA genes, and three non-coding control regions. The genome showed a slight A + T bias (A + T = 56.46%). Most PCGs were found to be located on the L-strand. Results of the phylogenetic analysis showed that N. stracheyi is closely related to N. hendersoni. Our results will help to clarify the phylogenetic relationship between Neolissochilus and Acrossocheilus species.
The genus Neolissochilus Rainboth, 1985 (Cypriniformes: Cyprinidae) includes 34 species of freshwater fish (Singh et al. Citation2021), which are widespread across tropical regions of southern to southeastern Asia (Rainboth Citation1991; Khaironizam et al. Citation2015). The classification of species from the Cyprinidae family remains unclear (Chen Citation2013). In China, fish from the genus Neolissochilus are commonly known as 'new smooth-lipped fish’ and their name has a meaning similar to that of Acrossocheilus, which means 'smooth-lipped fish' . The differentiation between Neolissochilus and Acrossocheilus species based only on morphology is ambiguous (Dasgupta Citation1988; Ambak and Jalal 2006; Sharma et al. Citation2019) (). Complete mitogenomes are being increasingly used to resolve ambiguity and provide accurate phylogenetic relationships between organisms. Preliminary studies have been performed on Neolissochilus species to analyze phylogenetics (Lalramliana et al. Citation2019; Gu et al. Citation2020; He et al. Citation2021); however, their phylogenetic analysis using mitogenome sequence has not been accomplished. In the present study, the complete mitochondrial genome of Neolissochilus hendersoni (Herre, 1940) was sequenced to analyze its phylogenetic position.
One specimen of N. hendersoni was collected from Daying River, Tengchong, Yunnan Province, China (24°36′36″ N, 97°49′12″ E) (). Part of the specimen was used in this study and the remaining sample was stored in the Biological Specimen Room of Zhejiang Ocean University (www.zjou.edu.cn, Yong-YaoGuo, [email protected]), voucher numeral LJL20211120. Total DNA from N. hendersoni sample was extracted using the E.Z.N. A® tissue DNA kit (Qiagen, Manchester, UK). The Illumina NovaSeq6000 platform (Illumina, San Diego, CA) was used to construct DNA libraries and to perform sequencing following protocols used by Zhang et al. (Citation2020). The raw sequencing data were filtered and 79,108,922 high-quality reads with a Q30% of 91.57% were obtained. The clean data were then assembled using Pilon v1.23. Gene annotation was performed using MITOS2 (http://mitos2.bioinf.uni-leipzig.de/index.py).
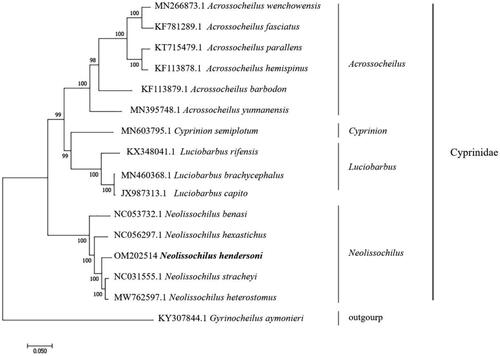
The complete mitogenome of N. hendersoni (GenBank accession no. OM202514) was found to be 16584 bp long. The Mitogenome contained two ribosomal RNA (12S rRNA and 16S rRNA) genes, 13 protein-coding genes (PCGs), 22 transfer RNA (tRNAs) genes, and three non-coding control regions. The overall base composition of N. hendersoni was A 31.79%, T 24.67%, G 15.78%, and C 27.76%. The sequence had a slight A + T bias with 56.46% A + T content. The maximum-likelihood (ML) phylogenetic tree was constructed using MEGA 7.0 with the GTR + G + I models (Xian et al. 2015) using the complete mitogenome of 16 fish species belonging to Acorssocheilus, Cyprinion, Luciobarbus, Neolissochilus, Gyrinocheilus, All complete mitogenome data are available for download from the Nucleotide at NCBI. As shown in the phylogenetic tree (), N. hendersoni is closely related to N. stracheyi. Further, the analysis indicates that the genus Neolissochilus is nearest in origin to the genus Luciobarbus and is farther from the genus Acrossocheilus in a phylogenetic position. These data will help clarify the phylogenetic relationship between Neolissochilus and Acrossocheilus species.
Ethical approval
This study was conducted following the guidelines and approval of the Institutional Animal Care and Use Committee at the Zhejiang Laboratory Animal Research Center and Zhejiang Ocean University. The approval number is 20210207.
Author contributions
Y.Guo and Y.C conceived the study. C.Yin and J.Wang collected the specimen. Y.Guo, Y.Wang, and C.Yin carried out the experiments and data analyses. Y.Guo wrote the manuscript with contributions from all authors. All authors approved the final manuscript and agreed to be accountable for all study aspects.
Acknowledgments
We thank the Taylor & Francis Editing Services (https://www.tandfeditingservices.com) for its linguistic assistance during the preparation of this manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data supporting this study’s findings are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/nuccore/OM202514.1/) under accession no. OM202514. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA816692, SRR18336527, and SAMN26688729.
Table 1. Difference in morphological features between Neolissochilus and Acrossocheilus.
Additional information
Funding
References
- Chen XY. 2013. Checklist of Fishes of Yunnan. Dongwuxue Yanjiu. 34(4):281–343.
- Dasgupta M. 1988. Fecundity of Acrossocheilus hexagonolepis (McClelland) from Garo Hills, Meghalaya. Uttar Pradesh Journal of Zoology 8(2):159–167.
- Gu W, Xu G, Huang T, Wang B. 2020. The complete mitochondrial genome of Neolissochilus benasi (Cypriniformes: Cyprinidae). Mitochondrial DNA Part B. 5(1):463–464.
- He J, Zhao C, Guo Y, Zhang H, Zhao B, Chu Z., 2021. Completely mitochondrial genome of Neolissochilus heterostomus. Mitochondrial DNA Part B. 6(9):2708–2709.
- Khaironizam MZ, Akaria-Ismail M, Armbruster JW. 2015. Cyprinid fishes of the genus Neolissochilus in Peninsular Malaysia. Zootaxa. 3962(1):139–157.
- Lalramliana n, Lalronunga S, Kumar S, Singh M. 2019. DNA barcoding revealed a new species of Neolissochilus Rainboth, 1985 from the Kaladan River of Mizoram, North East India. Mitochondrial DNA Part A. 30(1):52–59.
- Rainboth WJ. 1991. Cyprinids of Southeast Asia. In Cyprinid fishes. Springer, Dordrecht. p. 156–210.
- Sharma L, Ali S, Siva C, Kumar R, Barat A, Sahoo PK, Pande V. 2019. Genetic diversity and population structure of the threatened chocolate mahseer (Neolissochilus hexagonolepis McClelland 1839) based on SSR markers: implications for conservation management in Northeast India. Molecular Biology Reports. 46(5):5237–5249.
- Singh S, Mishra A, Pavan-Kumar A, Bidyasagar Singh S, Radhakrishnan KV, Nagpure NS, Chaudhari A., 2021. Characterization of the complete mitochondrial genome of Neolissochilus hexastichus (McClelland, 1839). Mitochondrial DNA Part B. 6(9):2461–2463.
- Zhang LJ, Wu L, Li Y, Li JG, Yang XK, Nie RE. 2020. The complete mitochondrial genome of the seed-borer weevil, Bruchidius uberatus (Coleoptera: Chrysomelidae: Bruchinae). Mitochondrial DNA Part B. 5(1):308–309.