Abstract
Astragalus melilotoides Pall. 1776 is a perennial leguminous forage, widely distributed in northern China, with cold, drought and disease resistance characteristics. Here, we determined the complete chloroplast (cp) genome sequence of A. melilotoides. It was 123,827 bp in length and 36.97% GC content with IR loss. The cp genome contained 110 complete genes, including 76 protein-coding genes, 30 tRNA genes, and four rRNA genes. A maximum likelihood (ML) phylogenetic tree revealed that A. melilotoides was related to A. americanus, A. gummifer, A. mongholicus, A. nakaianus, A. mongholicus var. nakaianus, and A. membranaceus var. membranaceus. The cp genome analysis of A. melilotoides will provide a reference for the phylogenetic study of Astragalus in the future.
Introduction
Astragalus melilotoides Pall. firstly discovered by Pall. in 1776 (https://www.tropicos.org/name/Search?name=Astragalus melilotoides), is a perennial leguminous forage widely distributed in desert steppe regions in northern China (Fu Citation1993). Due to its characteristics of drought-, cold-tolerance, water-holding capacity, disease resistance, and feeding value, A. melilotoides has been used in grassland improvement and forage supplementation (Fu Citation1993; Hou and Jia Citation2004). In order to improve the palatability, seed quality and yield of A. melilotoides, in the last decades, some scholars established a protoplast regeneration system to ensure its feeding quality (Hou and Jia Citation2004), and to explore the contribution of Agrobacterium rhizogenes to protoplasm regeneration (Zhang et al. Citation2008). The excellent characters of wild A. melilotoides can be cultivated by means of gene editing or somatic hybridization, these are crucially mediated by Agrobacterium rhizogenes (Zhang et al. Citation2008). Above studies laid a solid foundation for the feeding prospect of A. melilotoides. The value of A. melilotoides will be paid more and more attention in future, however, studies on the phylogeny of A. melilotoides remains limited. We reported the complete chloroplast (cp) genome sequence of A. melilotoides to provide reference for further study of the phylogenetic status in Fabaceae family.
Materials
The sample of A. melilotoides was collected from semi-fixed moving dune on southwest margin of Mu Us Sandy Land, Ningxia Hui Autonomous Region, China (38°4′39.04″N, 106°32′26.27″E, alt. 1163 m), and dried with silica gel. The specimen (No. 2022ASME001LY) was deposited at the herbarium of the Institute of Forestry and Grassland Ecology, Ningxia Academy of Agriculture and Forestry Science (http://www.nxaas.com.cn/, Wangsuo Liu, email: [email protected]). A. melilotoides is a common and excellent leguminous forage in the desert ecosystem, it is characterized by having white corolla, flag valve suborbicular, base with short stipe, wing valve slightly shorter than flag valve, apex unequal 2-lobed, base with short ear, keel valve shorter than wing valve, apex purplish, ovary subsessile, and glabrous (Fu Citation1993; ).
Methods
Genomic DNAs were extracted from samples by a modified CTAB method (Stefanova et al. Citation2013). The genome sequencing was performed by Illumina Hiseq 2500 at Biomarker Technologies Corporation. We assembled the cp genome via the program SPAdes3.11.0 (Nurk et al. Citation2013), with that of A. gummifer Labill. (1790) (GenBank: MN746310) as the initial reference. The cpDNA of A. melilotoides was annotated using Plann (Huang and Cronk Citation2015). The online tool OGDRAW (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html) was conducted to generate the cp genome maps (Greiner et al. Citation2019). To further clarify the A. melilotoides phylogenetic position, we downloaded the cp genome sequences of 34 Astragalus species in NCBI database and select two Oxytropis as outgroup. We aligned A. melilotoides and the other cp genomes by MAFFT-7.037 (Katoh and Standley Citation2013), and the maximum-likelihood tree was constructed by MEGA_X_10.2 with 1000 bootstrap replicates (Kumar et al. Citation2018). The sequence of A. melilotoides complete cp genome has been submitted to NCBI database (accession number ON854659).
Results and discussion
The complete cp genome of A. melilotoides was 123,827 bp in length, circular in form with IR loss, consistent with some of the species in Fabaceae (Ding et al. Citation2021; Guo et al. Citation2021; Ke et al. Citation2022). The cp genome encoded 110 genes, containing 76 protein-coding genes, 30 tRNA genes, and four rRNA genes. The overall GC content was 36.97% (). The maximum-likelihood phylogenetic tree showed that A. melilotoides was closely related to A. americanus, A. gummifer, A. mongholicus, A. nakaianus, A. mongholicus var. nakaianus, and A. membranaceus var. membranaceus (). Previous study for the phylogenetic characteristics of Astragalus were verified in the clustering tree in this study (Ding et al. Citation2021; Guo et al. Citation2021; Ke et al. Citation2022; Wang et al. Citation2016). The A. melilotoides complete cp genome provides valuable reference for the taxonomy and phylogenetic evolution study of Astragalus.
Figure 2. Gene map of A. melilotoides chloroplast genome. Genes shown inside the circle indicate that the direction of transcription is clockwise, while those shown outside are counterclockwise. Different groups of functional genes are indicated in different colors. The GC content is shown in the dashed area in the inner circle.
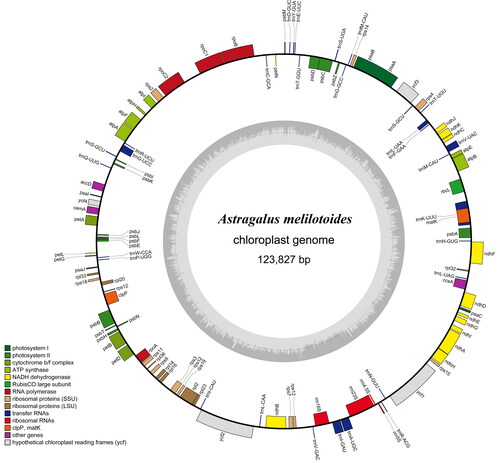
Figure 3. Phylogenetic relationship between Astragalus melilotoides and other 34 species of Astragalus based on the complete cp genome sequences from the NCBI database by using MEGA-X. Among this species, A. sinicus (OM287552) (Ke et al. Citation2022), A. galactites (MZ504977) (Ding et al. Citation2021), A. membranaceus var. membranaceus (KX255662) (Wang et al. Citation2016), A. laxmannii (MT786136) (Liu et al. Citation2020), and A. complanatus (MZ221114) (Yang Citation2021) have been published. Oxytropis arctobia and Oxytropis bicolor were chosen as the outgroups. The number on each node represents bootstrap values.

Conclusion
This study reported that the complete cp genome of A. melilotoides a typical circular form with IR loss, and 123,827 bp in length. A total of 110 genes were found in the cp genome of A. melilotoides, including 76 protein-coding genes, 30 tRNA genes, and 4 rRNA genes, and the GC content accounted for 36.97%. The phylogenetic tree reflects A. melilotoides has the closely relationships with A. americanus, A. gummifer, A. mongholicus, A. nakaianus, A. mongholicus var. nakaianus, and A. membranaceus var. membranaceus. This result provides a reference for the phylogenetic study of Astragalus in Fabaceae family.
Ethical approval
The collection and research process of plant material (A. melilotoides) was carried out in strict accordance with the guidelines provided by the herbarium of Ningxia Academy of Agriculture and Forestry Science and Chinese regulations.
Author contributions
WL planned and designed the research. YT and BJ sampled the plant materials, ZW, YT and BJ performed experiments, and WL analyzed the data. ZW and WL wrote the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI database at https://www.ncbi.nlm.nih.gov/, reference number ON854659. The associated BioProject, SRA, and Biosample numbers are PRJNA852531, SRR19939288, and SAMN29332762, respectively.
Additional information
Funding
References
- Ding X, Zhang C, Gao Y, Bei Z, Yan X. 2021. Characterization of the complete chloroplast genome of Astragalus galactites (Fabaceae). Mitochondrial DNA B Resour. 6(11):3278–3279.
- Fu K. 1993. Flora of China. Vol. 42. Beijing: Science Press.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Guo Y, Yao B, Yuan M, Li J. 2021. The complete chloroplast genome and phylogenetic analysis of Astragalus scaberrimus Bunge 1833. Mitochondrial DNA B Resour. 6(12):3364–3366.
- Hou SW, Jia JF. 2004. Plant regeneration from protoplasts isolated from embryogenic calli of the forage legume Astragalus melilotoides Pall. Plant Cell Rep. 22(10):741–746.
- Huang D, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Katoh K, Standley D. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Ke Q, Shangguan H, Liu W, Tang M, Bian J, Cui L, Li Y. 2022. The complete chloroplast genome and phylogenetic analysis of Astragalus sinicus Linne 1767. Mitochondrial DNA B Resour. 7(5):851–853.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Liu Y, Chen Y, Fu X. 2020. The complete chloroplast genome sequence of medicinal plant: Astragalus laxmannii (Fabaceae). Mitochondrial DNA B Resour. 5(3):3643–3644.
- Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Sirotkin Y, et al. 2013. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol. 20(10):714–737.
- Stefanova P, Taseva M, Georgieva T, Gotcheva V, Angelov A. 2013. A modified CTAB method for DNA extraction from soybean and meat products. Biotechnol Biotec Eq. 27(3):3803–3810.
- Wang B, Chen H, Ma H, Zhang H, Lei W, Wu W, Shao J, Jiang M, Zhang H, Jia Z, et al. 2016. Complete plastid genome of Astragalus membranaceus (Fisch.) Bunge var. membranaceus. Mitochondrial DNA B Resour. 1(1):517–519.
- Yang J. 2021. Characterization of the complete plastid genome of the perennial herb Astragalus complanatus Bunge (Fabales: fabaceae). Mitochondrial DNA B Resour. 6(12):3440–3442.
- Zhang GN, Jia JF, Hao JG, Wang XR, He T. 2008. Plant regeneration from mesophyll protoplasts of Agrobacterium rhizogenes-transformed Astragalus melilotoides. Biologia Plant. 52(2):373–376.

