Abstract
The mitochondrial genome (mitogenome) of Aleochara (Aleochara) curtula (Goeze, 1777) (Coleoptera: Staphylinidae) is reported. This mitogenome (GenBank accession no. OL675411) is 16,600 bp in size and consists of 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), and two ribosomal RNA genes (rRNA). Most PCGs use typical mitochondrial stop codon (TAR) except for cox3, which uses a single T residue. The A, G, T, and C nucleotide base composition of the mitogenome is 40.61%, 7.66%, 40.34%, and 11.39%, respectively. The phylogenetic analyses recovered the monophyly of Aleocharinae.
Keywords:
Staphylinidae is one of the largest coleopteran families with approximately 63,600 species (Irmler et al. Citation2018), and the subfamily Aleocharinae Fleming contains over 16,200 described species distributed worldwide (Leschen and Newton Citation2015). The aleocharine genus Aleochara Gravenhorst is recorded by over 400 species in 19 subgenera worldwide and 17 species in five subgenera in Korea (Ahn et al. Citation2017). Members of this genus are potential biocontrol agents against hygienic pests such as flies. Adults and larvae of the Aleochara species are ectoparasitoids of cyclorrhaphous Diptera, laying eggs near the fly larvae so that their offspring can trace the pupae (Klimaszewski Citation1984). In this study, we sequenced the complete mitochondrial genome of Aleochara (Aleochara) curtula (Goeze, 1777), a rove beetle species reported from regional outbreaks in the Republic of Korea and known to attack vacationers in the region when agitated (Lee Citation2019).
The specimens of A. curtula were collected in Cheongju-si, Chungcheongbuk-do, Republic of Korea (GPS coordinates: 36°37′N 127°27′E), which deposited at the Chungbuk National University Insect Collection (CBNUIC), Cheongju, Republic of Korea (voucher number: CBNU-202005-001; the person in charge of the collection: Jong-Seok Park, [email protected]). The total genomic DNA was extracted from an adult specimen of A. curtula using the MagAttract HMW DNA Kit (QIAGEN Biotech Co., Ltd) according to the manufacturer’s protocol. DNA library was prepared according to Illumina Truseq DNA PCR-free library preparation protocol and sequenced on the NovaSeq6000 sequencer (DNALink, Republic of Korea), using one Illumina lane. A total of 146,923,211 reads and 22,185,404,861 bp were generated. The mitochondrial genome sequence was assembled by GetOrganelle 1.6.4 (Jin et al. Citation2020) The mitogenome annotation was performed on the MITOS web server (Bernt et al. Citation2013) and adjusted with Geneious R10 (Biomatters, Auckland, New Zealand).
The total length of the complete mitochondrial genomes of A. curtula (GenBank accession no. OL675411) is 16,600 bp, which could be assembled into one circular contig. The A, G, T, and C nucleotide base composition of the mitogenome is 40.61%, 7.66%, 40.34%, and 11.39%, respectively. In general, the PCGs of A. curtula have an ATK start codon and a TAR stop codon sequence, with the exception of the nad1 gene, which uses the TTG start codon and the cox3 gene, which uses a stop codon with a single T residue.
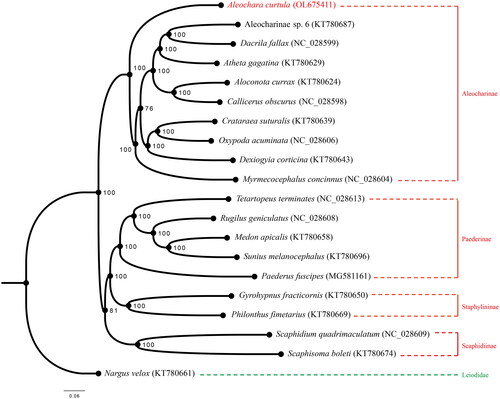
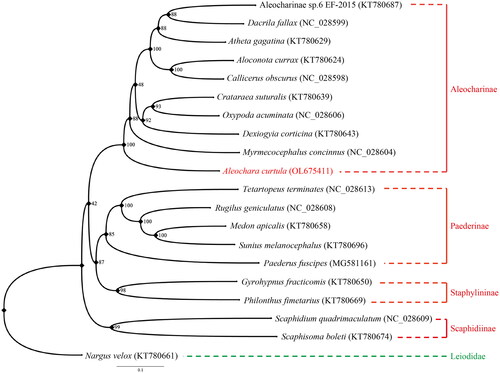
Maximum likelihood (ML) phylogenetic tree was reconstructed using IQ-TREE V.1.6.9 (Lin et al. Citation2018; Cai Citation2021) based on the 13 PCG nucleotide sequence data from our newly sequenced A. curtula mitogenome and 18 other species of Staphylinidae, together with one species of Leiodidae as an outgroup from GenBank (Mckenna et al. Citation2015; Lin et al. Citation2018; Cai et al. Citation2019; Kim et al. Citation2021). Each PCG was separately aligned using the L-INS-i algorithm of MAFFT 7.310 (Katoh and Toh Citation2008) and concatenated afterward for phylogenetic analysis. The best partitioning scheme and optimal nucleotide substitution models were determined within IQ-TREE: (1) GTR + F + I + G4 for nad2, cox1, cox3, nad3, nad4, nad4l and cytb; (2) GTR + F + R3 for cox2; (3) TVM + F + I + G4 for atp8; (4) TIM + F + I + G4 for atp6, nad5, nad6, and nad1. The standard bootstrap analysis was repeated 1,000 times to calculate bootstrap support values. Based on the ML tree reconstructed in this study, A. curtula is confirmed to constitute a monophyletic clade with other species of Aleocharinae (). Bayesian Inference was conducted by MrBayes 3.2.7 (Ronquist et al. Citation2012) with the same sequence alignment (). Subfamily Aleocharinae is sister to the other ingroup subfamilies, corresponding to BI phylogeny result inferred from the PCGs amino acid dataset ( of Song et al. Citation2021). Although some clades showed different relationships, both ML and BI results exhibited monophyly of each subfamily. It was also confirmed that mitogenomic data are valuable for analyzing phylogenetic relationships among the families of Coleoptera.
Figure 1. Maximum likelihood phylogeny of Staphylinidae based on the mitogenomic data of Aleochara (Aleochara) curtula and 19 other coleopteran species. Numbers on the nodes indicate bootstrap support values, and tip labels show taxon names with their respective GenBank accession numbers. Our newly sequenced A. curtula is marked in red, and the outgroup Leiodidae in green.
Author contributions
In this study, Chan-Jun Lee and Ji-Wook Kim analyzed the phylogeny and wrote the result part of the article. Jeesoo Yi, Yeon-Jae Choi, and Mi-Jeong Jeon collected specimens and wrote the introduction part. Sangil Kim analyzed the sequence data and polished the paper. Jong-Seok Park and Sung-Jin Cho contributed conceptualization of this study, funding acquisition, writing/revising of the manuscript, and approval of the final version. All authors have read and agree to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Mitochondrial genome sequence that supports the findings of this study is openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession number OL675411. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA820532, SRX14629784, and SAMN27014688 respectively. FAIRsharing DOI for data repositories is 10.25504/FAIRsharing.99sey6. All samples are stored in CBNUIC (Jong-Seok Park Ph.D, [email protected]). No applicable regulation or permission is needed for this study, which is related to any ethical issues.
Additional information
Funding
References
- Ahn KJ, Cho YB, Kim YH, Yoo IS, Newton AF. 2017. Checklist of the Staphylinidae (Coleoptera) in Korea. J Asia-Pac Biodivers. 10:279–336.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Cai CY, Wang YL, Liang L, Yin ZW, Thayer MK, Newton AF, Zhou YL. 2019. Congruence of morphological and molecular phylogenies of the rove beetle subfamily Staphylininae (Coleoptera: Staphylinidae). Sci Rep. 9:15137.
- Cai Y. 2021. The complete mitochondrial genome of Aleochara postica Walker, 1858 (Coleoptera: Staphylinidae). Mitochondrial DNA B Resour. 6(7):1994–1996.
- Irmler U, Klimaszewski J, Betz O. 2018. Introduction to the biology of rove beetles. In: Betz O, Irmler U, Klimaszewski J, editors, Biology of rove beetles (Staphylinidae): life history, evolution, ecology and distribution. 1st ed. Cham, Switzerland: Springer International Publishing; p. 1–4.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21:1–31.
- Katoh K, Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 9(4):286–298.
- Kim MJ, Chu M, Park JS, Kim SS, Kim I. 2021. Complete mitochondrial genome of the summer heath fritillary butterfly, Mellicta ambigua (Lepidoptera: Nymphalidae). Mitochondrial DNA B Resour. 6(5):1603–1605.
- Klimaszewski J. 1984. A revision of the genus Aleochara Gravenhorst of America north of Mexico (Coleoptera: Staphylinidae, Aleocharinae). Mem Ent Soc Can. 116:3–211.
- Lee HR. 2019. Occurrence on coastal resorts of Gyeongbuk East Sea… preventive emergency (in Korean). Intense 'Aleochara (Aleochara) curtula. [accessed 2021 Jul 9]. https://www.chosun.com/site/data/html_dir/2019/08/05/2019080500873.html.
- Leschen RAB, Newton AF. 2015. Checklist and type designations of New Zealand Aleocharinae (Coleoptera: Staphylinidae). Zootaxa. 4028(3):301–353.
- Lin A, Song N, Zhao X, Zhang F. 2018. Analysis of the nearly complete mitochondrial genome of Paederus fuscipes (Coleoptera: Staphylinidae). Mitochondrial DNA B Resour. 3(1):85–87.
- Mckenna DD, Farrell BD, Caterino MS, Farnum CW, Hawks DC, Maddison DR, Seago AE, Short AE, Newton AF, Thayer MK. 2015. Phylogeny and evolution of Staphyliniformia and Scarabaeiformia: forest litter as a stepping stone for diversification of nonphytophagous beetles. Syst Entomol. 40:35–60.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Song N, Zhai Q, Zhang Y. 2021. Higher-level phylogenetic relationships of rove beetles (Coleoptera, Staphylinidae) inferred from mitochondrial genome sequences. Mitochondrial DNA A DNA Mapp Seq Anal. 32:98–105.