Abstract
Here, we sequenced the complete mitogenome of Rhinogobius szechuanensis using the Illumina HiSeq platform and submitted the genome to Genbank with accession number OM617727. Assembly circular mitogenome (16,492 bp) consisted of 54.4% AT content, 13 protein-coding genes, 22 tRNA genes, two rRNA genes, an origin of light-strand replication, and a control region. Phylogenetic analysis supported that R. szechuanensis was grouped with R. rubromaculatus and clustered with other Rhinogobius species. The basal molecular data will be essential for further genetics studies such as evolution, taxonomy, DNA barcoding, and population genetics of Rhinogobius.
1. Introduction
Rhinogobius (Gill, 1859) is a genus widely distributed in the Western Pacific and East Asia, with approximately 129 species according to Fishbase statistics, and shows high species abundance and ecological diversity (Huang et al. Citation2016). In recent years, great advances have been made in resolving the phylogeny of Rhinogobius. Rhinogobius szechuanensis (Tchang, 1939), which belongs to the genus Rhinogobius within the subfamily Gobionellinae, is a freshwater goby endemic to the Minjiang River drainage of China. It is a warm temperate small-scale bottom fish, often inhabiting in flowing streams and rivers, and mostly moving among rocks and pebbles, and it feeds on small fish, aquatic insects, shrimps and crabs (Wu and Zhong, Citation2008). Because of its small size (∼ 70 mm) and unique body color, it is an ornamental fish popular in aquaria. It can be recognized by dorsal fin VI, I-8-9; anal fin I-7-8; pectoral fins 16–17; abdominal fin I-5; caudal fin 2 + 16 + 4; longitudinal scales 32–34; transverse scales 10–11; dorsal fin anterior scale 0; gill rake 9; no sensory canal; brown margin of each scale on the side of the body, forming reticular patterns (Wu and Zhong Citation2008) ().
Figure 1. Rhinogobius szechuanensis. Specimen was taken from the Minjiang River, Dujiangyan City, Sichuan Province, China. Photographs by Lin Song on 20 June 2021.

Rhinogobius szechuanensis has long been classified into the genus Glossogobius within the subfamily Gobiinae in the past based on morphological characteristics (Zhang Citation1996; Ye et al. Citation2015). Later, researchers proposed to classify it into the genus Rhinogobius within the subfamily Gobionellinae (Chen et al. Citation2008; Huang et al. Citation2016; Zhang and Zhao Citation2016). Up to present, only the 12S rRNA of R. szechuanensis was sequenced by Miya et al. (Citation2020), and still lacks strong molecular data to support whether it belongs to Rhinogobius. This research aimed to achieve the complete mitochondrial genome of R. szechuanensis to clarify its phylogenetic position. The mitogenome is useful as a reference for molecular species identification, and further genetics studies. Therefore, the research is of great significance.
2. Materials and methods
2.1. Sample collection and preservation
In this study, animals were collected under the permission of Jiangsu Agri-animal Husbandry Vocational College (NSF2021ZR14). Samples of R. szechuanensis were obtained from the Minjiang River, Dujiangyan City, Sichuan Province of China (31.0189° N, 103.5879° E) in June 2021. According to the morphological characteristics of R. szechuanensis provided by Wu and Zhang, (2008), the fish were identified by means of anatomical microscope and other tools. The samples were quickly frozen with liquid nitrogen and stored at −80 °C, and then transferred to the laboratory. The specimen was preserved in Aquatic Science and Technology Institution Herbarium (https://www.jsahvc.edu.cn/; Voucher number ASTIH-21b1108d26, Chen Xiao Jiang, [email protected]).
2.2. DNA extraction and sequencing
The total genomic DNA was extracted from a specimen muscle using the Tguide Cell/tissue genomic DNA Extraction Kit (OSR-M401) (Tiangen, Beijing, China). DNA samples were subjected to quality control: a) The concentration and purity of the samples were determined using NanoDrop 2000 (Thermo Fisher Scientific, USA), which required concentrations ≥20 ng/µL, total amount ≥ 100 ng, and OD260/OD280 = 1.8–2.2; b) The sample integrity was checked by agarose gel electrophoresis, which required that a major band of genomic DNA be visible and that no appreciable diffusion of degradation exists. The sequencing library was prepared by random fragmentation of the DNA sample, followed by PCR amplification, size selection and library quality check. The DNA raw reads were obtained from the sequencing of the constructed library on Illumina HiSeq 4000 Sequencing platform (Illumina, CA, USA). After the quality check process was conducted on FastQC Version 0.11.8 (Andrews Citation2015), the filtered reads were assembled into the complete mitogenome using MetaSPAdes with default parameters (Nurk et al. Citation2017), and the preliminary assembly results including maximum depth, minimum depth and average depth were 33,179 bp, 500 bp and 769.89 bp, respectively.
The resulting circular contig consensus sequence was annotated and verified with MITOS WebServer (http://mitos.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013). Organelle Genome Maps was drawn by OGDRAW (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html) (Greiner et al. Citation2019).
2.3. Phylogenetic analysis method
MEGA X was used to conduct the molecular phylogenetic (Kumar et al. Citation2018), and Maximum-likelihood (ML) method was applied to perform the phylogenetic analysis with 1000 bootstrap replicates. The phylogenetic tree based on 13 PCGs of R. szechuanensis and 22 published species of Gobionellinae, with Mugilogobius abei as outgroup. The best evolutionary model of the concatenated sequences was simulated to be mtREV + G + I + F for the lowest Bayesian information standard scores.
3. Results and discussion
3.1. Characteristics of R.s szechuanensis mitochondrial genome
The R. szechuanensis circular 16,492 bp mitogenome was composed of the following nucleotide base composition: 27.4% A, 27% T, 16.4% G, and 29.1% C, which demonstrated the AT-rich (54.4%) feature. The genome organization and transcriptional direction were identical to those of all known goby mitogenomes (Xie et al. Citation2015; Wang et al. Citation2019), containing 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, an origin light-strand replication (Ori-L, a small DNA region between tRNAAsn and tRNACys genes that form a stem-loop structure, which some studies have shown to be essential to start mtDNA light strand replication. It was determined according to the sequence blast comparison with the proximal species), and a control region (D-Loop). Among all 37 genes, ND6 and eight tRNA genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer(UCN), tRNAGlu, and tRNAPro) were encoded on the L-strand, the rest PCGs and tRNA genes were encoded on the H-strand (). Twelve mitochondrial PCGs for R. szechuanensis shared the regular initiation ATG, only the cytochrome c oxidase 1 (COX1) gene began with GTG, as found in other gobies (Yang et al. Citation2020; Tan et al. Citation2020). There were four different patterns of termination codons: TAA, TAG, T, and TA. The conserved 13 PCGs varied from 168 bp (ATP8) to 1839 bp (ND5). The lengths of 22 tRNAs ranged from 67 to 76 nucleotides. The large ribosomal gene (16S) was 1639 bp and the small (12S) was 950 bp, which was divided by tRNAVal. The 839 bp-long control regions were identified between tRNAPhe and tRNAPro ().
Figure 2. The complete mitochondrial genome map of R. szechuanensis (GenBank accession no. OM617727), consisting of 13 PCGs, 22 tRNAs, two rRNAs, the arrows represent direction of transcription. Genes encoded on H-strand and L-strand are displayed outside and inside the ring, respectively.
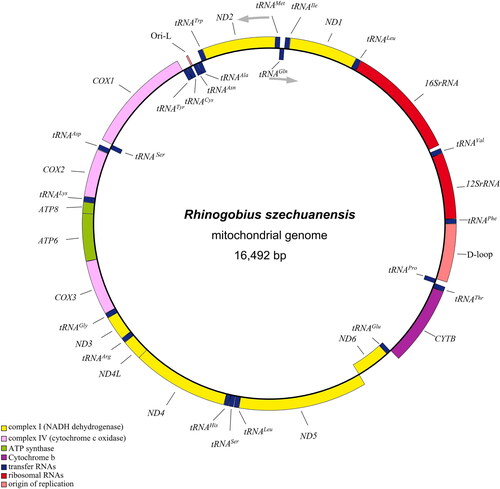
Table 1. Characteristics of the mitochondrial genome of R. szechuanensis.
3.2. Phylogenetic analysis
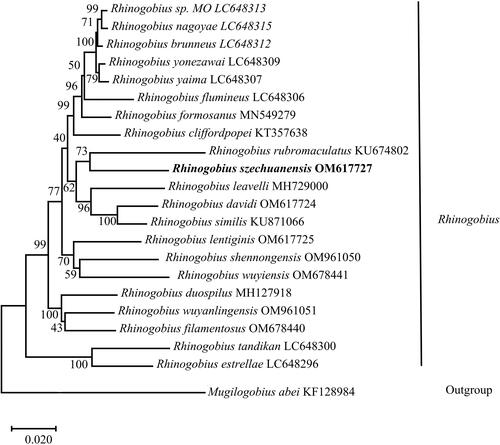
The phylogenetic position of R. szechuanensis was inferred by a phylogenetic tree using MEGA X based on the combined 13 PCGs for 23 Gobionellinae species (Cheng et al. Citation2005; Huang et al. Citation2013; Huang et al. Citation2015; Xie et al. Citation2015; Wang et al. Citation2019; Zhang and Shen Citation2019; Tan et al. Citation2020; Yang et al. Citation2020; Maeda et al. Citation2021). As shown in , the topological structure of the phylogenetic tree showed that R. szechuanensis and R. rubromaculatus gathered together, and they formed a sister group with the group include R. leavelli, Rhinogobius davidi, and Rhinogobius similis. The relationships among Rhinogobius were congruent with previous phylogenetic studies (Song et al. Citation2022). The results of phylogenetic analysis revealed the phylogenetic position of R. szechuanensis in the genus Rhinogobius at the molecular level. R. szechuanensis were clustered with all fish species of Rhinogobius, and within the genus R. szechuanensis and R. rubromaculatus were closely related.
Figure 3. Maximum-likelihood (ML) phylogenetic tree was reconstructed based on the concatenated 13 protein-coding genes of R. szechuanensis and other 22 Gobionellinae fishes. Accession numbers were indicated after the species names. Numbers at the nodes indicated bootstrap support values from 1000 replicates.
4. Conclusions
We reported the first complete mitochondrial genome assembly and annotation of R. szechuanensis using the next-generation sequencing technology. The circular mitogenome was 16,492 bp in length, contained 37genes encoding 13 PCGs, 22 tRNAs and two rRNAs. The phylogenetic position was inferred by a Maximum-likelihood phylogenetic tree based on the concatenated amino acid sequences of 23 Gobionellinae species, which supported that R. szechuanensis was grouped together with R. rubromaculatus, and clustered with other Rhinogobius species. In brief, molecular data revealed that it should belong to Rhinogobius, which justified without doubt the current classification status as correct and valid. The mitochondrial genomic data of R. szechuanensis provided in this study will aid future research on evolution, taxonomy, DNA barcoding and population genetics of Rhinogobius species.
Ethical statement
The experiments were approved by the Ethics Committee for Animal Experiments of Jiangsu Agri-animal Husbandry Vocational College, and conducted following the Chinese Association for the Laboratory Animal Sciences and the Institutional Animal Care and Use Committee (IACUC) protocols.
Author contributions
Wen Zhao Liu and Xiao Jiang Chen conceive and design the work, conduct the experiments, and drafting the paper, and Final approval of the version to be published; Lin Song and Hai Xia Liu were involved in the acquisition, analysis and interpretation of the data; the drafting of the paper. All authors agree to be accountable for all aspects of the work
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the reference number OM617727. The associated “BioProject”, “Bio-Sample” and “SRA” numbers are PRJNA808189, SAMN26035968, and SRR18065517 respectively.
Additional information
Funding
References
- Andrews S. 2015. FastQC: a quality control tool for high throughput sequence data [cited 2022 Jan 13]. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Cheng HL, Huang S, Lee SC. 2005. Morphological and molecular variation in Rhinogobius rubromaculatus (Pisces: gobiidae) in Taiwan. Zool Stud. 44:119–129.
- Chen IS, Cheng YH, Shao KT. 2008. A new species of Rhinogobius (Teleostei: gobiidae) from the Julongjiang basin in Fujian Province, China. Ichthyol Res. 55(4):335–343.
- Greiner S, Lehwark P, Bock R. 2019. Organellar genome draw (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Huang SP, Zeehan J, Chen I. 2013. A new genus of Hemigobius generic group goby based on morphological and molecular evidence, with description of a new species. J Mar Sci Technol. 21(7):19.
- Huang SP, Shen CN, Chen IS. 2015. The complete mitochondrial genome of the Abe’s mangrove goby Mugilogobius abei (Teleostei, Gobiidae). Mitochondrial DNA. 26(1):143–144.
- Huang SP, Chen IS, Shao KT. 2016. A new species of Rhinogobius (Teleostei: gobiidae) from Zhejiang Province, China. Ichthyol Res. 63(4):470–479.
- Huang SP, Chen IS, Jang-Liaw NH, Shao KT, Yung MMN. 2016. Complete mitochondrial reveals a new phylogenetic perspective on the brackish water goby mugilogobius group (teleostei: gobiidae: gobionellinae). Zoolog Sci. 33(5):566–574.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Maeda K, Shinzato C, Koyanagi R, Kunishima T, Kobayashi H, Satoh N, Palla HP. 2021. Two new species of Rhinogobius (Gobiiformes: Oxudercidae) from Palawan, Philippines, with their phylogenetic placement. Zootaxa. 5068(1):81–98.
- Miya M, Gotoh RO, Sado T. 2020. MiFish metabarcoding: a high-throughput approach for simultaneous detection of multiple fish species from environmental DNA and other samples. Fish Sci. 86(6):939–970. doi:10.1007/s12562-020-01461-x.
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. MetaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834.
- Song L, Chen XJ, Mao HX, Wang Q. 2022. Characterization and phylogenetic analysis of the complete mitochondrial genome of Rhinogobius wuyanlingensis (Gobiiformes: Gobiidae: Gobionellinae). Mitochondrial DNA Part B. 7(7):1323–1325.
- Tan HY, Yang YY, Zhang M, Chen XL. 2020. The complete mitochondrial genome of Rhinogobius duospilus (Gobiidae: Gobionellinae). Mitochondrial DNA Part B. 5(3):3406–3407.
- Wang D, Dai CX, Li Q, Li Y, Liu ZZ. 2019. Complete mitochondrial genome and phylogenic analysis of Rhinogobius cliffordpopei (Perciformes, Gobiidae). Mitochondrial DNA Part B. 4(2):2473–2474.
- Wu HL, Zhong JS. 2008. Fauna Sinica, Ostichthyes. Perciformes (V). Gobioidei. Beijing: Science Press (in Chinese)..
- Xie LP, Yang XF, Ma ZH, Yang RB. 2015. Complete mitochondrial genome of Rhinogobius giurinus (Perciformes: Gobiidae: Gobionellinae). Mitochondrial DNA. 26(2):321–322.
- Yang CJ, Chen Y, Chen Z, He GH, Zhong ZL, Xue WB. 2020. The next-generation sequencing reveals the complete mitochondrial genome of Rhinogobius formosanus (Perciformes: Gobiidae). Mitochondrial DNA B Resour. 5(3):2673–2674.
- Ye EQ, Xing YC, Zhang CG, Zhao YH. 2015. Catalogue of the type specimens in the fish collection of the National Zoological Museum, Institute of Zoology, Chinese Academy of Sciences, Beijing, China. Zootaxa. 3962:10–113.
- Zhang YF. 1996. Catalogue of the fish type specimens preserved in the fish collection of the Institute of Zoology, Academia Sinica. Acta Zootaxonomica Sinica. 21(4):498–503.
- Zhang CG, Zhao YH. 2016. Species diversity and distribution of inland fishes in China. Beijing, China: Science Press.
- Zhang FB, Shen YJ. 2019. Characterization of the complete mitochondrial genome of Rhinogobius leavelli (Perciformes: Gobiidae: Gobionellinae) and its phylogenetic analysis for Gobionellinae. Biologia. 74(5):493–499.
