Abstract
We provide the complete mitochondrial genome of the reef manta ray, Mobula alfredi, using an ezRAD approach. The total length of the mitogenome was 18,166 bp and contained 13 protein-coding genes, 22 transfer RNAs genes, two ribosomal RNA genes, and one non-coding control region. The gene organization and length are similar to other Mobula species. This reference mitogenome that includes the control region is expected to be a valuable resource for molecular-based species identification, population genomics, and phylogeography.
Introduction
Reef manta rays (Mobula alfredi, Krefft 1868) are emblematic inhabitants of coral reefs in tropical and subtropical oceans around the world (Last and Stevens Citation2009; Marshall et al. Citation2009). Many populations are in decline or may be threatened due to anthropogenic effects (Deakos et al. Citation2011; Croll et al. Citation2016; O’Malley et al. Citation2017; Stewart et al. Citation2018; Pate and Marshall Citation2020), which have contributed to their Vulnerable to Extinction status on the IUCN Red List of Threatened Species (Marshall et al. Citation2019). Despite their popularity and threatened status, our understanding of population structure remains limited. The whole mitogenome could be a powerful tool for population genomics of M. alfredi, as it offers high resolution assessment of gene flow and potential for exploring sex-based differences in behavior. However, no complete mitogenome reference is yet available for the species.
White et al. (Citation2017) reclassified the family Mobulidae using mitogenomes, exons and morphology into a monophyletic genus, Mobula, with 8 accepted species. This reclassification included the placement of the genus Manta in synonymy with the genus Mobula, which was supported by prior phylogenetic analysis (Poortvliet et al. Citation2015). White et al. (Citation2017) provided partial mitogenomes for most Mobulids, including M. alfredi (KX151653.1), however they were limited to the protein-coding genes and thus truncated before the control region. The control region (D-loop, displacement-loop) is non-coding and hypervariable and thus has value as an informative marker for evaluating gene flow across and population structure at fine spatial scales. Therefore, we sequenced the whole mitogenome of M. alfredi, including the control region, to provide a complete mitogenome reference valuable for application to species identification, population genomics and phylogeography of the reef manta ray.
Materials and methods
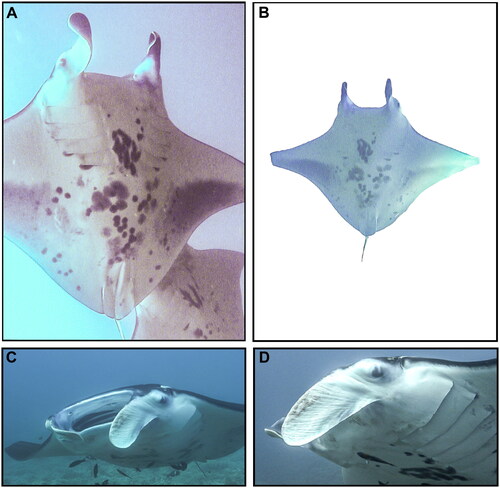
Tissue samples of several M. alfredi individuals were obtained on 25 November 2010 off Olowalu Reef, Maui, Hawai‘i, USA (20.7913°N, 156.5880°W), including from the individual pertinent to this study, an adult female reef manta ray known as ‘Bullseye’ (Catalog #176) in the Hawaiʻi Association for Marine Education and Research photo-identification catalog (www.hamerinhawaii.org). Voucher photographs are provided in . Biopsies (skin and muscle) were taken from the caudal end of the manta ray’s disk while on SCUBA using a modified Hawaiian sling containing a stainless-steel cylindrical biopsy tip (13 mm length, 5 mm diameter), preserved in 20% salt-saturated DMSO, and stored at −20 °C. Individuals are identified using unique ventral markings, gender and age-class are assigned based on clasper development in males (White et al. Citation2006; Marshall and Bennett Citation2010; Deakos et al. Citation2011) or mating scars and visible pregnancy in females (Marshall and Bennett Citation2010; Deakos Citation2012). Body size (disk width = 3.44 m) was measured using paired-laser photogrammetry as described in Deakos (Citation2010). Tissues are deposited at the Pacific Islands Fisheries Science Center (PIFSC-MOALF-HAMR176-B19; contact Jonathan Whitney, [email protected]).
Figure 1. Voucher photographs (A–D) of wild adult female Mobula alfredi (HAMER CatalogID#176 ‘Bullseye’) sampled from Maui Island, Hawai’i, USA in November 2010.
We extracted genomic DNA from tissue using an Omega E-Z 96 Tissue DNA Kit (Omega), following the manufacturer’s protocol. Due to the high prevalence of the GATC cut site in mitochondrial genomes as well as extensive random fragmentation in libraries, whole mitogenomes can be assembled from ezRAD libraries (Toonen et al. Citation2013; Tisthammer et al. Citation2016; Terraneo et al. Citation2018; Antaky et al. Citation2019), which simultaneously provide nuclear RAD loci for population genomics. We used the ezRAD approach (Toonen et al. Citation2013) to construct restriction-associated digest (RAD) reduced representation libraries with the enzyme DpnII (GATC cut site, New England Biolabs) following the ezRAD protocol (see Appendix 1 for detailed steps; Knapp et al. Citation2016). Genomic DNA was digested overnight with DpnII, end-repaired, 3′ ends adenylated, and ligated with Illumina TruSeq HT dual-indexed adapter sequences. DNA fragments from 300 to 425 bp (insert sizes 200–300 bp) were isolated using a PippenPrep (Sage Science). Adapter-ligated, size-selected fragments were amplified using PCR. Samples were cleaned using AMPureXP beads (Beckman-Coulter). DNA concentration was quantified using Accublue Quantitation (Biotium). A Bioanalyzer (Applied Biosystems) was used to check size-distribution of final amplified libraries. Libraries were normalized in equimolar concentrations (150 ng) and combined into pools with other libraries, bead cleaned and sequenced on one lane of an Illumina HiSeq 3000 (PE 150) at the UCLA Technology Center for Genomics and Bioinformatics. Raw reads were demultiplexed and only matched index pairs were retained.
We used the following workflow to assemble the M. alfredi mitogenome from ezRAD sequences. Raw reads were assessed using FastQC (Andrews Citation2010) and reads with adapters were filtered out using Cutadapt v1.11, (Martin Citation2011), and orphaned reads removed. We then used GetOrganelle v1.6.2d (Jin et al. Citation2020) to assemble mitogenomes de novo using the mitogenome of sister species M. birostris (KF413894 in Hinojosa-Alvarez et al. Citation2015) as the initial bait using all reads. The GetOrganelle assembly did not circularize but produced two long scaffolds totaling 18,097 bases. Scaffolds were aligned to 28 mitogenomes (4 complete, 24 partial) from 11 Mobula species available in GenBank () using MUSCLE in Geneious Prime 2020 (www.geneious.com). This alignment revealed a 69-base gap in the 16S rRNA gene in our draft assembly, which we initially replaced with Ns to join scaffolds into a single 18,166 base assembly. To inform this gap, we extracted fresh gDNA from the same individual using a DNeasy Blood & Tissue Kit (Qiagen) and amplified a 587-bp fragment of the 16S rRNA gene using primers 16Sar-L:5′-CGCCTGTTTATCAAAAACAT-3′ and 16Sbr-H:5′-CCGGTCTGAACTCAGATCACGT-3′ (Palumbi et al. Citation1991). PCRs were performed in 20 μL reactions of 10 μL Immomix Red (Bioline), 0.5 μL BSA, 1.0 μL of each primer (1 μM), 5.5 μL water and 2 μL of gDNA with the following conditions: 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, 50 °C for 30 s, 72 °C for 60 s, and a final extension of 72 °C for 10 min. Amplicons were purified with ExoSAP-IT (Thermo Fisher) and sequenced using an ABI 3730xl Sequencer (Applied Biosystems). The 16S sequences were aligned to the draft assembly using Geneious mapper with high sensitivity option. The 16S fragment was identical to overlapping sequence of the draft assembly and spanned the 69-base gap, generating a consensus sequence of the complete circular mitogenome (OP562409). The mitogenome was assembled from 27,434 paired Illumina reads and the 16S fragment, with mean coverage of 214× and high-quality base calls (% HQ) across 99.4% of the mitogenome. Gene annotation and validation of the circular mitogenome () was performed using MitoZ (Meng et al. Citation2019) and MitoAnnotator pipeline (Iwasaki et al. Citation2013). Pairwise sequence divergence was calculated between M. alfredi and sister species M. birostris (KF413894) using MEGA v.11 (Stecher et al. Citation2020, Tamura et al. Citation2021) on alignments of whole mitogenomes.
Figure 2. Map of the assembled Mobula alfredi mitochondrial genome (GenBank Accession: OP562409) consisting of 13 protein-coding genes (black), 22 transfer RNAs genes (red), two ribosomal RNA genes (light brown), and one non-coding control region (D-loop, dark brown). Genes encoded on the reverse strand and forward strand are illustrated inside the circle and outside the circle, respectively. The inner ring displays the GC content of the genome (every 5 bp), where the darker lines represent higher GC percent. This map was drawn using MitoAnnotator (Iwasaki et al. Citation2013).
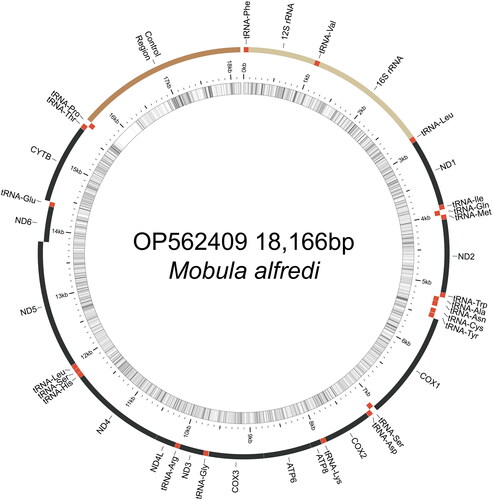
Table 1. Devil ray (Family Mobulidae) mitogenomes from Genbank.
Results
The mitochondrial genome of M. alfredi (OP562409) is estimated at 18,166 bases in length including 13 protein-coding genes, 22 transfer RNAs genes, two ribosomal RNA genes, and one non-coding control region (). Overall nucleotide composition was composed of 30.7% A, 29.8% T, 25.7% C, and 13.8% G. The gene organization and length are similar to other Mobula species, which range from 17,610 to 18,913 (). Mobula alfredi presents an AT-rich tandem repeat region in the control region, which varies in length and is found in other Mobulid rays (Hinojosa-Alvarez et al. Citation2015; Poortvliet et al. Citation2015; White et al. Citation2017). Pairwise sequence divergence between M. alfredi and sister species M. birostris was 0.009.
Discussion and conclusions
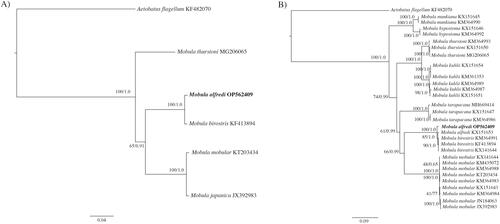
We present the first complete mitochondrial genome, including the control region, for Mobula alfredi. The complete genome phylogenetic tree () confirms the placement of OP562409 as sister to M. birostris and within the genus Mobula. The partial genome tree (excluding the control region) confirms the placement of OP562409 within the M. alfredi species clade (). The divergence observed in the mitogenome between M. alfredi and M. birostris (d = 0.009) is similar to interspecific divergence rates that are suitable for species delineation among Mobula species (White et al. Citation2017). This complete circular reference mitogenome from the Hawaiian Islands includes the control region and is expected to be valuable for molecular-based species identification, population genomics, and phylogeography.
Figure 3. Molecular phylogenetic reconstruction of Mobula. Presented are rooted Bayesian trees based on (A) all available congeneric (n = 4) complete mitochondrial genomes (i.e. including the control region), and (B) all congeneric (n = 28) partial mitochondrial genomes (i.e. excluding the control region) (). Alignment and branch support analyses were performed in Geneious Prime v. 2022.0.1 The best fit sequence evolution model for both datasets, GTR + G (gamma = 0.2670, complete genome; gamma = 0.2650, partial genome), was identified by the Akaike Information Criterion using jModelTest v. 2.1 (Guindon and Gascuel Citation2003; Darriba et al. Citation2012). Phylogenetic reconstruction using a maximum likelihood (ML) analysis was created using PHYML v.3.0.1 (Guindon et al. Citation2010) as implemented in Geneious Prime with clade support assessed with 1000 non-parametric bootstrap replicates. Bayesian inference (BI) analysis was run using MrBayes v.2.2.4 (Huelsenbeck and Ronquist Citation2001; Ronquist and Huelsenbeck Citation2003) by running a pair of independent searches for 1 million generations, with trees saved every 1000 generations and the first 250 trees discarded as burn-in. Sequence divergence is represented on the scale bar. Branch support is presented as ML/BI. Aetobatus flagellum was selected as the outgroup. The resulting trees confirm the placement of OP562409 with M. alfredi, as sister to M. birostris and reinforces phylogenetic relationships established by previous studies (Poortvliet et al. Citation2015; White et al. Citation2017).
Ethical approval
Part of this research was conducted under the University of Hawai‘i Animal Care & Use Committee, Protocol No. 08-591-2, and Assurance number A3423-01. This study complied with the International Union for Conservation of Nature (IUCN) policies for research involving species at risk of extinction, the Convention on Biological Diversity and the Convention on the Trade in Endangered Species of Wild Fauna and Flora. We used non-lethal sampling methods, and no animal was caught, handled, or removed from its natural habitat for the purpose of this study.
Author contributions
JW, RC, and MD conceived, designed, and implemented the study. MD performed all fieldwork and sampling. JW performed all benchwork. JW and RC performed all bioinformatics and analyzed the data. JW wrote the manuscript with RC and MD.
Supplemental Material
Download MS Word (22.6 KB)Acknowledgements
Special thanks to Lee James with Ultimate Whale Watch Adventure, Craig Venema, and Amy Miller for providing a boat and valuable time in the field. Thanks to Stephen Karl, Brian Bowen, Rob Toonen, and Jamison Gove for their support of this research. Thanks to the State of Hawai‘i Division of Aquatic Resources. Thanks to Nan Himmelsbach and Andrea Schmidt for assistance with 16S sequencing.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under the accession no. OP562409. The associated BioProject, SRA, and BioSample accession numbers are PRJNA899543, SRR22233901, and SAMN31097093 respectively.
Additional information
Funding
References
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- Antaky CC, Kitamura PK, Knapp IS, Toonen RJ, Price MR. 2019. The complete mitochondrial genome of the band-rumped Storm Petrel (Oceanodroma castro). Mitochondrial DNA Part B. 4(1):1271–1272.
- Aschliman NC, Nishida M, Miya M, Inoue JG, Rosana KM, Naylor GJP. 2012. Body plan convergence in the evolution of skates and rays (Chondrichthyes: Batoidea). Mol Phylogenet Evol. 63(1):28–42.
- Bustamante C, Barría C, Vargas-Caro C, Ovenden JR, Bennett MB. 2016. The phylogenetic position of the giant devil ray Mobula mobular (Bonnaterre, 1788) (Myliobatiformes, Myliobatidae) inferred from the mitochondrial genome. Mitochondrial DNA Part A. 27(5):3540–3541.
- Chandrasekaran K, Dhinakarasamy I, Jayavel S, Rajendran T, Bhoopathy S, Gopal D, Ramalingam K, Ajmal Khan S. 2022. Complete sequence and characterization of the Mobula tarapacana (Sicklefin Devilray) mitochondrial genome and its phylogenetic implications. Journal of King Saud University - Science. 34(3):101909.doi:10.1016/j.jksus.2022.101909.
- Croll DA, Dewar H, Dulvy NK, Fernando D, Francis MP, Galván‐Magaña F, Hall M, Heinrichs S, Marshall A, Mccauley D, et al. 2016. Vulnerabilities and fisheries impacts: the uncertain future of manta and devil rays. Aquatic Conserv Mar Freshw Ecosyst. 26(3):562–575.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772–772.
- Deakos M, Baker J, Bejder L. 2011. Characteristics of a manta ray Manta alfredi population off Maui, Hawaii, and implications for management. Mar Ecol Prog Ser. 429:245–260.
- Deakos MH. 2010. Paired-laser photogrammetry as a simple and accurate system for measuring the body size of free-ranging manta rays Manta alfredi. Aquat Biol. 10(1):1–10.
- Deakos MH. 2012. The reproductive ecology of resident manta rays (Manta alfredi) off Maui, Hawaii, with an emphasis on body size. Environ Biol Fish. 94(2):443–456.
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321.
- Guindon S, Gascuel O. 2003. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst Biol. 5:696–704.
- Hinojosa-Alvarez S, Díaz-Jaimes P, Marcet-Houben M, Gabaldón T. 2015. The complete mitochondrial genome of the Giant Manta ray, Manta birostris. Mitochondrial DNA. 26(5):787–788.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30(11):2531–2540.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Knapp I, Puritz J, Bird C, Whitney J, Sudek M, Forsman Z, Toonen R. 2016. ezRAD- an accessible next-generation RAD sequencing protocol suitable for non-model organisms_v3.2. Protocols.io. doi:10.17504/protocols.io.e9pbh5n.
- Last P, Stevens J. 2009. Sharks and rays of Australia. 2nd ed. Cambridge (MA): Harvard University Press.
- Marshall A, Barreto R, Carlson J, Fernando D, Fordham S, Francis M, Herman K, Jabado R, Liu K, Pacoureau N, et al. 2019. Mobula alfredi. The IUCN red list of threatened species 2019: e.T195459A68632178. doi:10.2305/IUCN.UK.2019-3.RLTS.T195459A68632178.en.
- Marshall AD, Bennett MB. 2010. Reproductive ecology of the reef manta ray Manta alfredi in southern Mozambique. J Fish Biol. 77(1):169–190.
- Marshall AD, Compagno LJ, Bennett MB. 2009. Redescription of the genus Manta with resurrection of Manta alfredi. Zootaxa. 2301(1):1–28.
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17(1):10–13.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- O’Malley M p, Townsend KA, Hilton P, Heinrichs S, Stewart JD. 2017. Characterization of the trade in manta and devil ray gill plates in China and South‐east Asia through trader surveys. Aquatic Conserv Mar Freshw Ecosyst. 27(2):394–413.
- Palumbi S, Martin A, Romano S, McMillan W, Stice L, Grabowski G. 1991. A simple fool’s guide to PCR, v2.0. Honolulu: University of Hawaii Department of Zoology and Kewalo Marine Laboratory, Special Publication; p. 1–23.
- Pate J, Marshall A. 2020. Urban manta rays: potential manta ray nursery habitat along a highly developed Florida coastline. Endang Species Res. 43:51–64.
- Poortvliet M, Hoarau G. 2013. The complete mitochondrial genome of the Spinetail Devilray, Mobula japanica. Mitochondrial DNA. 24(1):28–30.
- Poortvliet M, Olsen JL, Croll DA, Bernardi G, Newton K, Kollias S, O’Sullivan J, Fernando D, Stevens G, Galván Magaña F, et al. 2015. A dated molecular phylogeny of manta and devil rays (Mobulidae) based on mitogenome and nuclear sequences. Mol Phylogenet Evol. 83:72–85.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Santillán-Lugo B, Llera-Herrera R, Corro-Espinosa D, Oñate-González EC, Rodríguez-Domínguez G, Saavedra-Sotelo NC. 2017. Complete mitochondrial genome of the Devil Ray, Mobula thurstoni (Lloyd, 1908) (Myliobatiformes: Myliobatidae). Mitochondrial DNA Part B. 2: 868–870.
- Stecher G, Tamura K, Kumar S. 2020. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol Biol Evol. 37(4):1237–1239.
- Stewart JD, Jaine FRA, Armstrong AJ, Armstrong AO, Bennett MB, Burgess KB, Couturier LIE, Croll DA, Cronin MR, Deakos MH, et al. 2018. Research priorities to support effective manta and devil ray conservation. Front Mar Sci. 5:314.
- Tamura K, Stecher G, Kumar S. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 38(7):3022–3027.
- Terraneo TI, Arrigoni R, Benzoni F, Forsman ZH, Berumen ML. 2018. Using ezRAD to reconstruct the complete mitochondrial genome of Porites fontanesii (Cnidaria: Scleractinia). Mitochondrial DNA B Resour. 3(1):173–174.
- Tisthammer KH, Forsman ZH, Sindorf VL, Massey TL, Bielecki CR, Toonen RJ. 2016. The complete mitochondrial genome of the lobe coral Porites lobata (Anthozoa: Scleractinia) sequenced using ezRAD. Mitochondrial DNA B Resour. 1(1):247–249.
- Toonen RJ, Puritz JB, Forsman ZH, Whitney JL, Fernandez-Silva I, Andrews KR, Bird CE. 2013. ezRAD: a simplified method for genomic genotyping in non-model organisms. PeerJ. 1:e203.
- White WT, Corrigan S, Yang L, Henderson A, Bazinet A, Swofford D, Naylor G. 2017. Phylogeny of the manta and devilrays (Chondrichthyes: Mobulidae), with an updated taxonomic arrangement for the family. Zool J Linn Soc. 182(1):50–75.
- White WT, Giles J, Potter IC, Dharmadi . 2006. Data on the bycatch fishery and reproductive biology of mobulid rays (Myliobatiformes) in Indonesia. Fisheries Res. 82(1-3):65–73.
