Abstract
The Great Marquis or Bassarona dunya is a butterfly species commonly found in the tropical regions of Asia, America, and Africa. This butterfly is a member of the subfamily Limenitidinae and the classification within this subfamily has been unstable. Here, we report the first complete mitochondrial genome (mitogenome) of B. dunya sampled from Malaysia. The mitogenome is 15,242 bp long, comprising a set of 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), two ribosomal RNAs (rRNAs), and an A + T rich region. All PCGs were initiated by the typical ATN codon, except for COX1 which started with a CGA start codon. Nine PCGs were terminated with a TAA or TAG stop codon, while COX1, COX2, NAD4, and NAD5 ended with an incomplete T. The 12S and 16S rRNAs were 716 bp and 1269 bp in length, respectively. Phylogenetic analysis supported the placement of B. dunya within Limenitidinae with a high support value.
Introduction
Bassarona dunya (Doubleday, 1848) is a butterfly from the subfamily Limenitidinae and is distributed mainly in the tropical regions of Asia, America, and Africa (Hui-Yun et al. Citation2022). They are commonly identified based on the uniform pale brown, with a postdiscal series of creamy white spots on both wings (Corbet et al. Citation2021) (). This subfamily has been subjected to a long history of complex taxonomic classification, and recent phylogenetic studies based on molecular data have gradually unraveled their taxonomic position both at the subfamily and lower classification level (Dhungel and Wahlberg Citation2018; Wu et al. Citation2019; Wahlberg et al. Citation2020; Hui-Yun et al. Citation2022; Liu et al. Citation2022). To date, a complete mitogenome of B. dunya has yet to be reported publicly. In this study, we sequenced and analyzed the first complete mitogenome of B. dunya from Malaysia which will become an important resource in further addressing the taxonomic issues of this subfamily.
Materials and methods
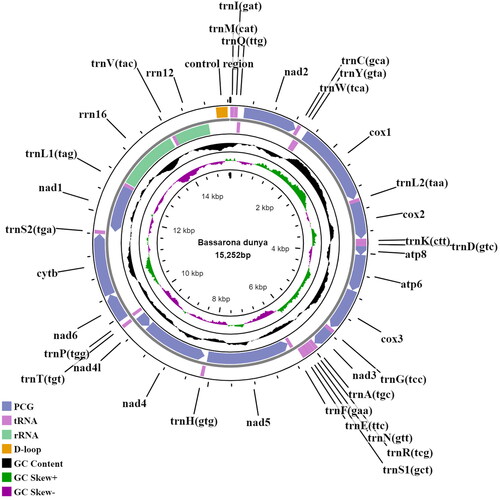
The adult B. dunya was collected from Ayer Hitam Forest Reserve Johor, Malaysia (2.025680, 102.794151). The specimen was deposited in Universiti Tun Hussein Onn Malaysia (UTHM) (https://uthm.edu.my; Dr Aqilah Awg Abdul Rahman; [email protected]) with the voucher ID DIB031. Total genomic DNA was extracted from the hind leg tissue using the Qiagen Blood and Tissue Kit (Qiagen, Valencia, CA) following the manufacturer’s instruction, and fragmented via a Bioruptor® system. For library preparation, NEBNext® Ultra™ II DNA Library Prep Kit for Illumina® was used prior to sequencing by Illumina NovaSeq 6000 system (PE150). The raw reads obtained were pre-processed using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and then trimmed for sequencing adapters, as well as low quality reads using AdapterRemoval (Schubert et al. Citation2016). For the mitogenome assembly, NOVOPlasty v.4.2 (Dierckxsens et al. Citation2017) was used, with a seed reference (BOLD ID: YB-KHC6757) before running through a PALEOMIX BAM pipeline (Schubert et al. Citation2014) (default parameters) to remove reads shorter than 15 bp after trimming (Miga et al. Citation2022). A total of 10,447,566 reads were retained and annotated using MITOS2 web server (Bernt et al. Citation2013). The predicted protein-coding genes (PCGs) were further verified via ORFFinder (https://www.ncbi.nlm.nih.gov/orffinder/) to improve the annotation. Tablet (Milne et al. Citation2010) was also used to visualize the assembled mitogenome for indels and sequence coverage. A mitogenome map of the sequenced B. dunya in this work was generated using Proksee (https://proksee.ca/), an updated version of the CGView web server (Grant and Stothard Citation2008) as displayed in .
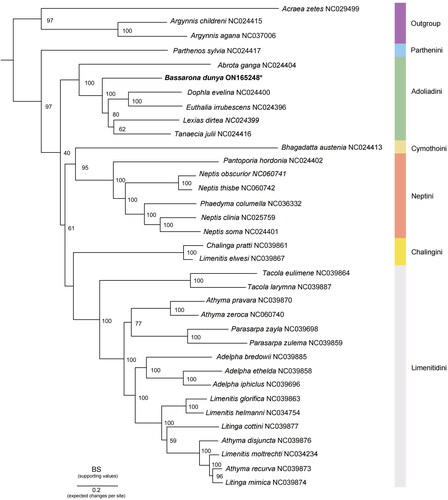
To investigate the phylogenetic relationships within this subfamily, 13 concatenated PCGs from 35 Nymphalidae mitogenome sequences were used to reconstruct the maximum-likelihood (ML) tree. As we focus on the subfamily Limenitidinae, the combined dataset comprises three Heliconiinae and 32 Limenitidinae, including four recognized tribes (Dhungel and Wahlberg Citation2018). The mitogenomes of Acraea zetes (NC029499), Argynnis sagana (NC037006), and Argynnis childreni (NC024415) from the subfamily Heliconiinae were used as the outgroups. Phylogenetic analysis was performed using IQ-Tree (Nguyen et al. Citation2015), under an edge-linked partition model with 5000 ultrafast bootstrap replicates. The best partitioning schemes and evolutionary model were determined by PartitionFinder v2.2.1 (Lanfear et al. Citation2017) ().
Figure 3. Phylogenetic analysis of B. dunya sequenced in this study, as indicated with an asterisk, and other 34 Nymphalidae mitogenomes. The node values represent the bootstrap of ultrafast 5000 replicates (BS), whereas each color indicates the tribes in Limenitidinae. Outgroup: Acraea zetes (NC029499) (Timmermans et al. Citation2016); Argynnis sagana (Liu et al. Citation2018); Argynnis childreni (NC024415) (Wu et al. 2014). Ingroup: Abrota ganga (NC024404), Euthalia irrubescens (NC024396), Lexias dirtea (NC024399), Dophla evelina (NC024400), Tanaecia julii (NC024416), Bhagadatta austenia (NC024413), Parthenos sylvia (NC024417), Neptis soma (NC024401), and Pantoporia hordonia (NC024402) (Wu et al. 2014); Adelpha bredowii (NC039885), Adelpha ethelda (NC039858), Adelpha iphiclus (NC039696), Athyma pravara (NC039870), Athyma recurva (NC039873), Athyma disjuncta (NC039876), Limenitis glorifica (NC039863), Litinga mimica (NC039874), Litinga cottini (NC039877), Parasarpa zulema (NC039859), Parasarpa zayla (NC039698), Tacola larymna (NC039887), Tacola eulimene (NC039864), Chalinga pratti (NC039861), and Limenitis elwesi (NC039867) (Wu et al. Citation2019); Athyma zeroca (NC060740), Neptis obscurior (NC060741), and Neptis thisbe (NC060742) (Liu et al. Citation2021); Neptis clinia (NC025759) (Tang, et al. Citation2014); Phaedyma columella (NC036332) (Chen et al. Citation2018); unpublished: Limenitis helmanni (NC034754) and Limenitis moltrechti (NC034234); this study: Bassarona dunya (ON165248).
Results
The complete mitogenome of B. dunya (GenBank accession no.: ON165248) is 15,242 bp in length, encoding 13 PCGs, 22 transfer RNAs (tRNAs), two ribosomal RNAs (rRNAs), and a control region. The mitogenome has a depth coverage of 167X and is A + T biased (81.45%). It has a nucleotide composition of A (38.51%), T (42.93%), TC (10.96%), and G (7.59%). The PCGs have a total length of 11,226 bp, whereas the tRNAs are 1461 bp, ranging from 61 bp (tRNA-Ser) to 71 bp (tRNA-Lys). The length of the 12S and 16S rRNA genes are 716 bp and 1269 bp, respectively. Out of the 13 PCGs, 12 were initiated by the typical ATN start codon, while COX1 utilizes the CGA codon. Four PCGs (COX1, COX2, NAD4, and NAD5) ended with an incomplete stop codon, while others were terminated either by TAA or TAG stop codon. Additionally, BLASTn analysis (McGinnis and Madden Citation2004) was done on the COX1 sequence of this species which showed 97.04% similar to the B. dunya COX1 sequence in GenBank (accession no.: GQ864742).
Phylogenetic analysis showed that all tribes formed monophyletic clades. The ML tree recovered the relationship of Parthenini+(Adoliadini+((Cymothoini + Neptini)+(Chalingini + Limetidini))). The analysis also showed that the newly sequenced B. dunya formed a clade with the other Asian Adoliadini genera (Dhungel and Wahlberg Citation2018; Hui-Yun et al. Citation2022) and is sister to Abrota ganga (NC024404) (Wu et al. Citation2014). The placement of B. dunya within the clade is also highly supported (100%).
Discussion and conclusions
This study provided the complete mitogenome of B. dunya and was deposited in GenBank (accession no.: ON165248). The newly sequenced B. dunya reported shared similar gene characteristics with the other genera in Limenitidinae. It has the shortest mitogenome length compared to other genera in the Adoliadini tribe, but it is still within the range of the typical Lepidoptera mitogenome. These differences could be influenced by the length of the control region (Cheng et al. Citation2018). The utilization of the CGA start codon is also common in Lepidoptera mitogenome and the phenomena of the incomplete stop codon is presumed to be associated with the polyadenylation processes (Chen et al. Citation2020). For phylogenetic analysis, the monophyly of Adoliadini is strongly supported (BS = 100), and the position of Parthenini as sister to the other Limenitidinae tribes is consistent with other reported studies (Wahlberg et al. Citation2020; Hui-Yun et al. Citation2022; Liu et al. Citation2022). The mitogenome of B. dunya sequenced in this study provides useful information on the phylogeny of the Adoliadini tribe, and to further address their taxonomic issues, extensive taxon sampling should be considered.
Author contributions
Marylin Miga: conceptualization, methodology, data curation, software, and writing – original draft preparation. Yap Yong Zi: data curation, methodology, data validation, reviewing, and editing. Puteri Nur Syahzanani Jahari: conceptualization, methodology, software, data validation, reviewing, and editing. Sivachandran Parimannan and Heera Rajandas: formal analysis, resources, and funding acquisition. Muhammad Abu Bakar-Latiff and Yap Jing Wei: methodology and funding acquisition. Mohd Shahir Shamsir: methodology. Faezah Mohd Salleh: conceptualization, methodology, resources, reviewing and editing, supervision and funding acquisition. All authors have read and approved the manuscript.
Acknowledgements
This work was done with the permission by the Forestry Department of Peninsular Malaysia under the Research permit JH/100 Jld. 30 (18).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. ON165248. The associated BioProject, SRA, and BioSample numbers are PRJNA753627, SRR15422669, and SAMN20720549, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chen YC, Wang CT, Lees DC, Wu LW. 2018. Higher DNA insert fragment sizes improve mitogenomic assemblies from metagenomic pyrosequencing datasets: an example using Limenitidinae butterflies (Lepidoptera, Nymphalidae). Mitochondrial DNA A DNA Mapp Seq Anal. 29(6):840–845.
- Chen L, Wahlberg N, Liao CQ, Wang CB, Ma FZ, Huang GH. 2020. Fourteen complete mitochondrial genomes of butterflies from the genus Lethe (Lepidoptera, Nymphalidae, Satyrinae) with mitogenome-based phylogenetic analysis. Genomics. 112(6):4435–4441.
- Cheng YC, Chen MY, Wang JF, Liang AP, Lin CP. 2018. Some mitochondrial genes perform better for damselfly phylogenetics: species- and population-level analyses of four complete mitogenomes of Euphaea sibling species. Syst Entomol. 43(4):702–715.
- Corbet A, Pendlebury H, Poorten G, van der Poorten N. 2021. The butterflies of the Malay Peninsula. 5th ed. (2020). Revised by George Michael van der Poorten and Nancy van der Poorten. Southdene Sdn. Bhd.
- Dhungel B, Wahlberg N. 2018. Molecular systematics of the subfamily Limenitidinae (Lepidoptera: Nymphalidae). PeerJ. 6:e4311.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Grant JR, Stothard P. 2008. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36(Web Server Issue):181–184.
- Hui-Yun T, Chiba H, Lohman DJ, Yen SH, Aduse-Poku K, Ohshima Y, Wu LW. 2022. Out of Asia: intercontinental dispersals after the Eocene-Oligocene transition shaped the zoogeography of Limenitidinae butterflies (Lepidoptera: Nymphalidae). Mol Phylogenet Evol. 170(February):107444.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. Partitionfinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773.
- Liu N, Fang L, Zhang Y. 2021. The Complete Mitochondrial Genomes of Four Species in the Subfamily Limenitidinae (Lepidoptera, Nymphalidae) and a Phylogenetic Analysis. Insects. 13(1):16.
- Liu N, Li N, Yang P, Sun C, Fang J, Wang S. 2018. The complete mitochondrial genome of Damora sagana and phylogenetic analyses of the family Nymphalidae. Genes Genom. 40(1):109–122.
- Liu N, Fang L, Zhang Y. 2022. The complete mitochondrial genomes of four species in the subfamily Limenitidinae (Lepidoptera, Nymphalidae) and a phylogenetic analysis. Insects. 13:16.
- McGinnis S, Madden TL. 2004. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 32(Web Server Issue):20–25.
- Miga M, Jahari PNS, Vei Siang C, Kamarudin KR, Shamsir MS, Tokiman L, Parimannan S, Rajandas H, Mohamed F, Salleh FM. 2022. The complete mitochondrial genome data of the Common Rose butterfly, Pachliopta aristolochiae (Lepidoptera, Papilionoidea, Papilionidae) from Malaysia. Data Brief. 40:107740.
- Milne I, Bayer M, Cardle L, Shaw P, Stephen G, Wright F, Marshall D. 2010. Tablet-next generation sequence assembly visualization. Bioinformatics. 26(3):401–402.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Schubert M, Ermini L, Sarkissian CD, Jónsson H, Ginolhac A, Schaefer R, Martin MD, Fernández R, Kircher M, McCue M, et al. 2014. Characterization of ancient and modern genomes by SNP detection and phylogenomic and metagenomic analysis using PALEOMIX. Nat Protoc. 9(5):1056–1082.
- Schubert M, Lindgreen S, Orlando L. 2016. AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res Notes. 9(88):88.
- Tang M, Tan M, Meng G, Yang S, Su X, Liu S, Song W, Li Y, Wu Q, Zhang A, et al. 2014. Multiplex sequencing of pooled mitochondrial genomes—a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 42(22):e166.
- Timmermans MJ, Lees DC, Thompson MJ, Sáfián S, Brattström O. 2016. Mitogenomics of ‘Old World Acraea’ butterflies reveals a highly divergent ‘Bematistes’. Mol Phylogenet Evol. 97:233–241.
- Wahlberg N, Maresova J, Murillo-Ramos L, Collins S, Wu LW. 2020. The phylogenetic positions of Bhagadatta Moore, 1898, Kumothales Overlaet, 1940 and Harmilla Aurivillius, 1892 (Lepidoptera, Nymphalidae, Limenitidinae) based on molecular data. Nota Lepidopterol. 43(2018):167–171.
- Wu L-W, Lin L-H, Lees DC, Hsu Y-F. 2014. Mitogenomic sequences effectively recover relationships within brush-footed butterflies (Lepidoptera: Nymphalidae). BMC Genomics. 15(1):468.
- Wu LW, Chiba H, Lees DC, Ohshima Y, Jeng ML. 2019. Unravelling relationships among the shared stripes of sailors: mitogenomic phylogeny of Limenitidini butterflies (Lepidoptera, Nymphalidae, Limenitidinae), focusing on the genera Athyma and Limenitis. Mol Phylogenet Evol. 130(August):60–66.