Abstract
Castanopsis fordii Hance 1884 is a typical evergreen broad-leaved forest plant in the south subtropical and middle subtropical regions of China. It has high utilization value in wood production and soil erosion protection. Here, we first reported and characterized the complete chloroplast (cp) genome sequence of C. fordii based on Illumina paired-end sequencing data. The complete cp genome sequence of C. fordii was 160,853 base pairs (bp) in length which contained two inverted repeats (IRs) of 25,699 bp separated by a large single-copy (LSC) and a small single copy (SSC) of 90,474 bp and 18,981 bp, respectively. The cpDNA contained 129 genes, comprising 85 protein-coding genes, 36 tRNA genes, 8 rRNA genes. The overall GC content of the plastome was 36.8%. Phylogenetic analysis base on 14 chloroplast genomes indicated that C. fordii was closely related to the species C. tibetana and C. concinna in Fagaceae.
Introduction
Castanopsis fordii belongs to the genus Castanopsis of Fagaceae family is naturally distributed in southeastern China with the Nanling Mountains as the distribution center, and is often the main tree species constituting the local evergreen broad-leaved forest (Lian Bei et al. Citation2018). C. fordii with straight trunk, fast growth and reddish-brown heartwood is a common wood species in southern China and has high economic value (Xu et al. Citation2010). In this study, we reported the complete chloroplast (cp) genome of C. fordii based on Illumina pair-end sequencing data, which was helpful for well understanding its characteristics and the origin in evolution.
Materials and methods
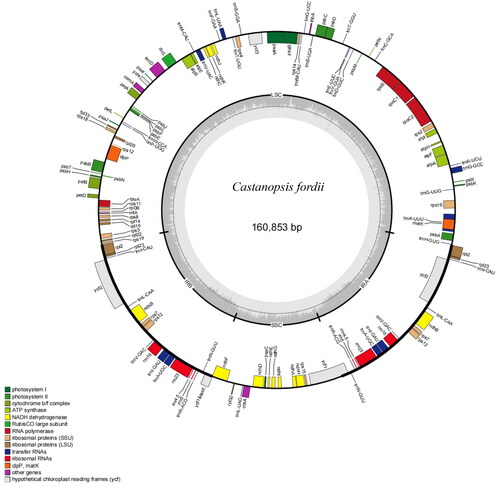
Fresh leaves of C. fordii (, Yong-lin Huang, [email protected]) were collected from the Haiyang Mountain nature reserve in yangshuo of Guangxi province, China (24°59′ N, 110°55′ E). The voucher specimen has been deposited in the Guangxi Key Laboratory of Plant Functional Phytochemicals and Sustainable Utilization, Guangxi Institute of Botany (http://www.gxib.cn/, Yong-lin Huang, [email protected]) under the voucher number MZ 20211223. DNA extraction from fresh leaf tissue was randomly disrupted into 300–500 bp by the Covaris ultrasonic breaker for library construction. The constructed library was sequenced using Illumina NovaSeq6000 platform, yielding approximately 2GB data. These trimmed reads were assembled by NOVOPlasty version 4.2 (Dierckxsens et al. Citation2016). The assembled genome was annotated using PGA (https://github.com/quxiaojian/PGA) (Qu et al. Citation2019) and Geseq with default settings (https://chlorobox.mpimp-golm.mpg.de/geseq.html) (Tillich et al. Citation2017). The annotation result was drawn with the online tool OGDRAW default parameters (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html) (Greiner et al. Citation2019) ().
Results
The complete cp genome sequence of C. fordii was 160,853 base pairs (bp) () in length which contained two inverted repeats (IRs) of 25,699 bp separated by a large single-copy (LSC) and a small single copy (SSC) of 90,474 bp and 18,981 bp, respectively. The cpDNA contained 129 genes, comprising 85 protein-coding genes, 36 tRNA genes, 8 rRNA genes. Most of the genes occurred in a single copy; however, six protein-coding genes (ndhB, rpl2, rpl23, rps12, rps7 and ycf2), seven tRNA genes (trnl-CAU, trnV-GAC, trnR-ACG, trnL-CAA, trnl-GAU, trnN-GUU and trnA-UGC), and four rRNA genes (4.5S, 5S, 16S and 23S) were totally duplicated. The over all GC content of the plastome was 36.8%, and the corresponding values in LSC was 35.07%. No gene has not been predicted. The annotated chloroplast genome of C. fordii has been deposited in GenBank with accession number ON710841.
Figure 2. The Genome map of C. fordii. Genes inside the circle are transcribed clockwise, and those on the outside are transcribed counter-clockwise. Genes are colored according to functional categories. The darker grey area in the inner circle corresponds to GC content, whereas the lighter grey corresponds to AT content.
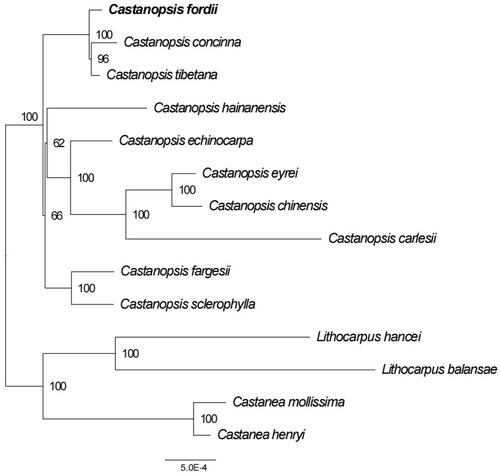
In order to reveal the phylogenetic position of C. fordii with other members of Fagaceae, a phylogenetic analysis was performed by MAFFT v7.158b (Katoh and Standley Citation2013) based on 11 complete cp genomes of Fagaceae, and Lithocarpus hancei, Lithocarpus balansae as outgroups. Then, the phylogenetic tree with 1000 ultrafast bootstrap (UFBoot) replicates (Minh et al. Citation2013) was constructed by RAxML. As shown in the ML phylogenetic tree (), the genus Castanea and Lithocarpus formed a monophyletic clade with high bootstrap value, respectively. C. fordii was closely related to C. concinna and C. tibetana. These results based on complete cp genome reported here will lay a basis for the study of phylogeny, phylogeography and population genetic diversity of C. fordii.
Figure 3. The ML phylogenetic tree based on fourteen complete chloroplast genomes. Maximum-likelihood phylogenetic tree was constructed by GTRCATI Model of RAxML based on complete cp genomes. The sequence type is the nucleotide sequence of the conservative coding gene. The bootstrap check value is 100. Numbers close to each node are bootstrap support values. Accession number: Castanopsis concinna KT793041.1 (Shi et al. Citation2022), Castanopsis hainanensis MG383644.1 (Ogoma et al. Citation2022), Castanopsis fargesii NC_047230.1 (Li et al. Citation2022), Castanopsis carlesii NC_057119.1 (Wang et al. Citation2022), Castanopsis sclerophylla MT627605.1 (Miao et al. Citation2020), Castanopsis echinocarpa NC_023801.1 (Guo et al. Citation2022), Castanopsis tibetana ON710842 (Sanchez-Puerta and Abbona Citation2014), Castanopsis eyrei ON550006, Castanopsis chinensis ON500675, Castanopsis fordii ON710841 (Li et al. Citation2022), Lithocarpus balansae KP299291.1 (Shi et al. Citation2022), Castanea henryi KX954615.1 (Shi et al. Citation2022), Castanea mollissima NC_014674 (Jansen et al. Citation2011), Lithocarpus hancei MW375417.1 (Shi et al. Citation2022). Castanopsis eyrei and Castanopsis chinensis have no publications and can only cite the sequences by accession numbers.
Discussion and conclusion
The chloroplast genome structures of the Fagaceae species that have been studied all contain one LSC region one SSC region and two IR regions, which is consistent with the basic structural characteristics of angiosperm chloroplast genomes. At the same time, it was found that the variation in the LSC region accounted for the vast majority of the variation in the whole genome, and the difference in the length of the whole genome was mainly caused by the difference in the length of the LSC. In this study, a phylogenetic tree was constructed based on the complete cp genomes sequences of 14 Fagaceae species. The classification of each species was basically consistent with the traditional taxonomy. This study lays the foundation for further studies on the evolution of Fagaceae genomes.
Author contributions
Ya-feng Wang, Yonglin Huang and Rui-jie He were involved in the conception and design. Ya-feng Wang was involved in the drafting of the paper. Yonglin Huang Zhang-bin Liu and Bing-yuan Yang make critical revisions to intellectual content. Yonglin Huang and Zhang-bin Liu were involved in the final approval of the version to be published; and that all authors agree to be accountable for all aspects of the work.
Sampling statement
Our materials are not listed in the Wild Plants Under State Protection in China, so we need not any permission or license to collect them; and we only sampled a few leaf materials for molecular experiment and didn’t damage any plant.
Disclosure statement
No potential conflict of interest was reported by the authors. The collection of plant material was carried out in accordance with national or international regulations.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession no. ON710841. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA864116, SRR20726142, and SAMN30074217 respectively.”
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):gkw955.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Guo S, Liao X, Chen S, Liao B, Guo Y, Cheng R, Xiao S, Hu H, Chen J, Pei J, et al. 2022. A comparative analysis of the chloroplast genomes of four Polygonum medicinal plants. Front Genet. 13:764534.
- Jansen RK, Saski C, Lee S-B, Hansen AK, Daniell H. 2011. Complete plastid genome sequences of three Rosids (Castanea, Prunus, Theobroma): evidence for at least two independent transfers of rpl22 to the nucleus. Mol Biol Evol. 28(1):835–847.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Li Y, Wang TR, Kozlowski G, Liu M, Yi L, Song Y. 2022. Complete chloroplast genome of an endangered species Quercus litseoides, and its comparative, evolutionary, and phylogenetic study with other Quercus Section Cyclobalanopsis Species. Genes (Basel). 13(7):1184.
- Lian Bei W, Xiao Chun S, Li Na Z, Dan Ping L, Jian Fei L, De Xiang Z. 2018. Relationship between diameter structure and interspecies of Castanopsis fordii at Yunzhong Mountain in Anxi. J Southwest Forestry Univ. 38(05):116–123.
- Miao J, Wang Y, Zhang Y, Tong L, Tang G, Xu X.,. 2020. The complete chloroplast genome sequence of Castanopsis sclerophylla (Lindl.) Schott (Fagaceae). Mitochondrial DNA B Resour. 5(3):2848–2849.
- Minh BQ, Nguyen MAT, Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195.
- Ogoma CA, Liu J, Stull GW, Wambulwa MC, Oyebanji O, Milne RI, Monro AK, Zhao Y, Li D-Z, Wu Z-Y, et al. 2022. Deep insights into the plastome evolution and phylogenetic relationships of the Tribe Urticeae (Family Urticaceae). Front Plant Sci. 13:870949.
- Qu XJ, Moore MJ, Li DZ, Yi T-S. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12.
- Sanchez-Puerta MV, Abbona CC. 2014. The chloroplast genome of Hyoscyamus niger and a phylogenetic study of the tribe Hyoscyameae (Solanaceae). PLoS One. 9(5):e98353.
- Shi X, Xu W, Wan M, Sun Q, Chen Q, Zhao C, Sun K, Shu Y. 2022. Comparative analysis of chloroplast genomes of three medicinal Carpesium species: genome structures and phylogenetic relationships. PLoS One. 17(8):e0272563.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wang J, Qian J, Jiang Y, Chen X, Zheng B, Chen S, Yang F, Xu Z, Duan B. 2022. Comparative analysis of chloroplast genome and new insights into phylogenetic relationships of polygonatum and Tribe Polygonateae. Front Plant Sci. 13:882189.
- Xu YF, Cao FX, Yu XL, Chengjing QI. 2010. Analysis on community structures and spatial patterns of Castanopsis fordii in Nanling Region of Hunan. Forest Res Manag. 8(04):22–26.

