Abstract
The mitochondrial genome of Cuspidaria undata (Verrill, 1884) was sequenced in full using Illumina HiSeq 2500. The circular mitochondrial DNA (mtDNA) was 16,266 bp in size, encoded 37 genes, and contained 13 protein-coding genes (PCGs), 2 rRNAs and 22 tRNAs. The gene order of the 13 PCGs in this species exhibited extensive rearrangement and differences in comparison to other Cuspidariidae, indicating that gene order is not conserved within this family. Phylogenetic analysis based on 13 PCGs and 2 rRNAs recovered a monophyletic Cuspidariidae.
The family Cuspidariidae Dall, 1886. belongs to the class Bivalvia, within the superorder Anomalodesmata (Worms Citation2020), and includes 19 genera and 254 species (Morton and Machado Citation2019). Cuspidariidae species are deep-sea predatory bivalves and are the most diversified group among the septibranch bivalves (Amano and Kurita Citation2020). Deep water mollusks have been sparsely studied and only one complete mitogenome of Cuspidariidae has previously been sequenced, one which belongs to the genus Tropidomya Dall & Smith, 1886 (Williams et al. Citation2017). In this study, the complete mitochondrial genome of Cuspidaria undata (Verrill, 1884) was sequenced to understand the genomic structure and phylogenetic relationships within this family ().
A specimen of C. undata was collected from the Southern Indian Ocean (16.95110S, 89.66835E) during the China Ocean 52th voyage. The sample (Accession No. FIO2018015207; Contact person: Zhou Zheng, [email protected]) was stored at −80 °C at the Key Laboratory of Marine Eco-Environmental Science and Technology, First Institute of Oceanography, Ministry of Natural Resources. Total DNA was extracted from muscle tissue using a DNeasy Blood & Tissue DNA kit (QIAGEN) and thereafter sequenced using the Illumina HiSeq 2500 Sequencing Platform (Beijing, China). The mitochondrial genome was assembled using NOVOplasty (Dierckxsens et al. Citation2017) and annotated using the MITOS Web Server (Bernt et al. Citation2013). Bandage (Wick et al. Citation2015) was used to verify the circular structure of the mitochondrial genome. The complete mitogenome of C. undata was submitted to GenBank, registration number ON360998.1.
The mitochondrial genome of C. undata was 16,266 bp in length, and contained 13 PCGs, 2 rRNAs and 22 tRNAs. The mtDNA of C. undata composition was 26.1% A, 10.4% C, 21.4% G, and 42.1% T. The percentage of A + T with the C. undata mtDNA was 68.2%. The 22 tRNA-coding genes ranged in size from 60 bp through to 74 bp. The gene order of the 13 PCGs was COX1, NAD6, COB, NAD4L, COX2, ATP6, NAD2, NAD4, ATP8, NAD1, NAD5, NAD3 and COX3, which exhibited extensive rearrangement in comparison to the gene order previously published Tropidomya abbreviata (Forbes, 1843), also within Cuspidariidae (Williams et al. Citation2017), indicating that the mitochondrial gene order of this family is not conservative. In addition, we made a table describing the characteristics of C. undata mitochondrial genome (), and drew the mitochondrial genome map of C. undata () using OrganellarGenomeDRAW (OGDRAW) version 1.3.1 (Greiner et al. Citation2019).
Figure 1. The maximum likelihood tree of 10 bivalves based on 13 PCGs and 2 rRNAs. Number at branch represents bootstrap probability. The following sequences were used: KX815961.1 (Williams et al. Citation2017), KX815958.1 (Williams et al. Citation2017), KX815962.1 (Williams et al. Citation2017), KX815963.1 (Williams et al. Citation2017), KF534717.1 (Park and Ahn Citation2015), KX815957.1 (Williams et al. Citation2017), KX815960.1 (Williams et al. Citation2017), KR873102.1 (Wu et al. Citation2016), MG385135.1 (Soroka and Burzyński Citation2017).
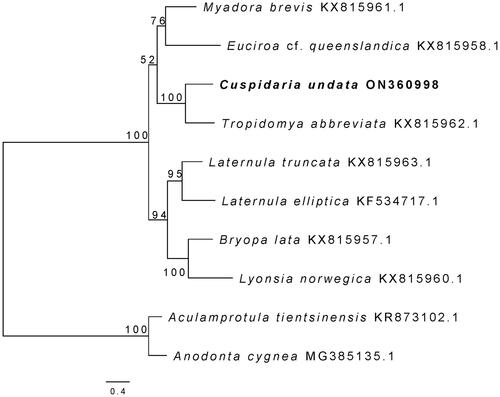
Figure 2. Map of the mitochondrial genome of C. undata. The mitochondrial genome of C. undata was 16,266 bp in length, and contained 13 PCGs, 2 rRNAs and 22 tRNAs, marked in different colors in the map.
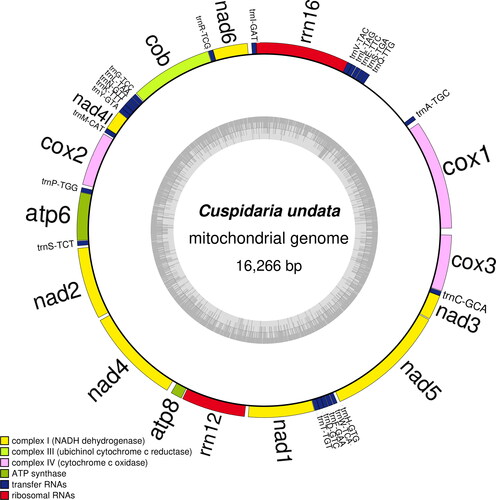
Table 1. Mitochondrial genome of C. undata with gene order, positions, lengths, nucleic acid composition, coding strand, start and stop codons, anticodons and amino acid size.
The maximum likelihood (ML) phylogenetic tree of C. undata was constructed based on the 13 PCGs and 2 rRNAs, containing seven other Anomalodesmata species and two Palaeoheterodonta species as outgroups. Nucleotide sequences from each mitogene were aligned using MAFFT v7.313 (Katoh et al. Citation2002) and then trimmed with Gblocks 0.9 b (Talavera and Castresana Citation2007). All of the aligned gene sets were concatenated using SequenceMatrix v1.7.8 (Vaidya et al. Citation2011). The best fitting nucleotide-substitution model of each partition was evaluated using PartitionFinder2 (Lanfear et al. Citation2017). ML analysis was analyzed using IQ-TREE v. 1.6.8 (Nguyen et al. Citation2015) with 1,000 bootstrap replicates. The phylogenetic position of C. undata (ON360998.1) was sister to T. abbreviata and the family Cuspidariodea recovered a monophyletic clade.
Ethical approval
In this study, all experimental protocols relating to animal experiments were in accordance with the measures for the Administration of Affairs Concerning Experimental Animal of Shandong Province, China (approved by the Shandong Provincial Government in 1992). All animal experiments in this study were approved by the Ethical Committee of Ministry of Natural Resources of the People’s Republic of China. All studies were conducted in accordance with ethical guidelines and the legal requirements of the study country.
Author contributions
Yating Bao, Meiling Ge and Yaoyao Zhao were involved in the conception and design, analysis and interpretation of the data. Yating Bao and Meiling Ge drafted the paper. Qinzeng Xu participated in the formal analysis and data curation of the project, and revised it critically for intellectual content. Zhou Zheng provided the funds and resource for this research and gave final approval of the version to be published. All authors agree to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/nuccore/ON360998) (GeneBank Number: ON360998.1). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA848421, SRR19647706, and SAMN29005581, respectively.
Additional information
Funding
References
- Amano K, Kurita H. 2020. A new Paleocene species of Myonera (Bivalvia: Cuspidariidae) from eastern Hokkaido, Northern Japan. Nautilus Greenville Then Sanibel. 134(1):51–56.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773.
- Morton B, Machado FM. 2019. Predatory marine bivalves: a review. Adv Mar Biol. 84:1–98.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Park H, Ahn DH. 2015. Complete mitochondrial genome of the Antarctic soft-shelled clam, Laternula elliptica (Bivalvia; Laternulidae). Mitochondrial DNA. 26(4):642–643.
- Soroka M, Burzyński A. 2017. Hermaphroditic freshwater mussel Anodonta cygnea does not have supranumerary open reading frames in the mitogenome. Mitochondrial DNA B Resour. 2(2):862–864.
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577.
- Vaidya G, Lohman DJ, Meier R. 2011. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 27(2):171–180.
- Williams ST, Foster PG, Hughes C, Harper EM, Taylor JD, Littlewood DTJ, Dyal P, Hopkins KP, Briscoe AG. 2017. Curious bivalves: systematic utility and unusual properties of anomalodesmatan mitochondrial genomes. Mol Phylogenet Evol. 110:60–72.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352.
- Worms. 2020. World register of marine species. http://www.marinespecies.org/aphia.php?p=taxdetails&id=1788.
- Wu RW, An CT, Wu XP, Zhou CH, Ouyang S. 2016. Complete maternal mitochondrial genome of freshwater mussel Aculamprotula tientsinensis (Bivalvia: unionidae: unioninae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(6):4520–4521.
