Abstract
Rubus irritans Focke is a type of tonifying kidney-essence herb used in China. We present the complete chloroplast (cp) genome of R. irritans, a member of the genus Rubus. The complete cp genome of R. irritans was 155,286 bp long and consisted of an 84,613 bp long large single-copy (LSC) region, an 18,697 bp long SSC region, and a pair of 25,988 bp long inverted repeats (IR). Furthermore, the plastid genome contained 130 genes, including 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The overall GC content of the genome was 37.29%. Based on the complete cp genome, phylogenetic analysis revealed that R. irritans is closely related to R. amabilis.
Introduction
According to the Flora of China (Editorial Committee of Flora of China, Chinese Academy of Sciences Citation1985), R. irritans Focke 1910 belongs to the Rosaceae family and is found in 27 Chinese provinces (Li et al. Citation2000). It is a dwarf shrub (about 10–60 cm tall) with pubescent and glandular hairs and branches covered in purple-red needles. Rubus irritans is a valuable plant resource for both medicine and food. According to the Compendium of Materia Medica, its stems, roots, leaves, and seeds can be used as medicine. We present here the complete cp genome of R. irritans, which is expected to improve Rosaceae utilization and phylogenetic investigation.
Materials
Rubus irritans leaves were collected from a mature plant in the General Ditch, Qinghai Province, China (34°29′4.30′′N, 102°08′19.10′′E). This study was approved by the Qinghai Academy of Agricultural and Forestry Sciences and complied with National Wild Plant Protective Regulations. The specimen was deposited at the herbarium of the pharmacy department of Qinghai University in Xining, China (https://yxy.qhu.edu.cn/, contact person: ShiBing Yang; Email: [email protected]) under the voucher number 630224190519002LY.
Methods
The modified CTBA method was used to extract genomic DNA (Doyle Citation1987). The complete chloroplast genome was sequenced on the Illumina HiSeq2500 platform (Illumina, USA), assembled with SPAdes 3.10.1 (Bankevich et al. Citation2012), and annotated with CPGAVAS2 (Shi et al. Citation2019). MAFFT was used to perform multiple sequence alignment on the sequences. The GTR and the mountain climbing model were chosen, and 1,000 bootstrap repeats were inferred using the maximum likelihood (ML) method with RaxML-HPC2 on the CIPRES web server’s TG ver7.2.8 (Stamatakis et al. Citation2008). As the best ML alternative model, the general time-reversible model was chosen.
Results
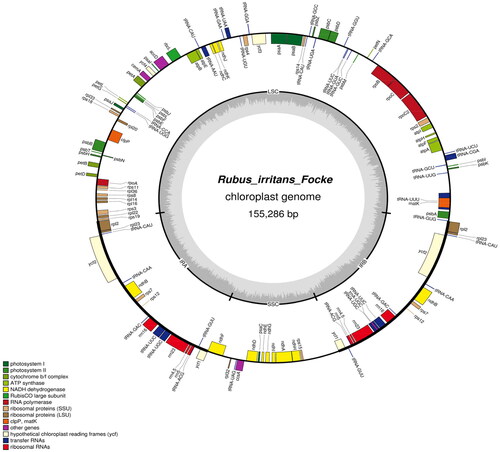
The R. irritans (MN652919) cp genome sequence was 155,286 bp along. A pair of IRs of 25,988 bp, an LSC region of 84,613 bp, and an SSC region of 18,697 bp comprised the typical quadripartite structure. The chloroplast genome had a GC content of 37.29%. There were 130 predicted genes, 85 of which were protein-coding genes, 37 of which were tRNA genes, and 8 of which were rRNA genes. For the cpSSR analysis, we used MISA V1.0, which found 229 SSR microsatellites and 149, 9, 62, 7, and 2 mono-, di-, tri-, tetra-, and penta-nucleotide repeats, respectively. The richest SSRs were mononucleotide SSRs (65.07%). There were 24,896 codons in the protein-coding genes. The two most abundant amino acids, were leucine and isoleucine, which each had 2,652 codons (approximately 10.65% of the total) and 2,102 codons (approximately 8.44% of the total). and depict the entire plant and the Genome Map of R. irritans, respectively.
Figure 1. The morphological characteristics of R. irritans. Photos A and B show the entire plant and its growing environment, respectively (photos taken by Shibing Yang in General Ditch, Qinghai Province).

Figure 2. Genome map of R. irritans. The forward-encoding and reverse-encoding genes are located on the inner and outer sides of the circle, respectively.
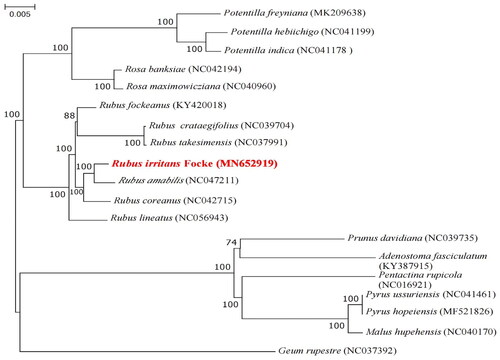
Based on the complete cp genome phylogenetic analysis, R. irritans is closely related to R. amabilis (). The complete cp sequences were obtained from the NCBI (www.ncbi.nlm.nih.gov).
Figure 3. Phylogenomic tree of R. irritans (in red) and 18 species constructed using ML method based on complete cp genome sequences. Numbers in the nodes were bootstrap values from 1000 replicates. The following sequences were used: Potentilla freyniana (Park et al. Citation2019), Potentilla hebiichigo, Potentilla indica (Liu et al. Citation2021), Rosa banksiae, Rosa maximowicziana (Kim et al. Citation2019), Rubus fockeanus, Rubus crataegifolius, Rubus takesimensis (Liu et al. Citation2021), Rubus amabilis, Rubus coreanus (Zhang et al. Citation2021), Rubus lineatus (Lim et al. Citation2021), Prunus davidiana (Wang et al. Citation2020), Adenostoma fasciculatum KY387915 (Kim and Kim Citation2016), Pyrus ussuriensis, Pyrus hopeiensis (Cho et al. Citation2019), Malus hupehensis (Li et al. Citation2020), Geum rupestre (Zhang et al. Citation2022).
Discussion and conclusion
According to the findings of this study, R. irritans is closely related to R. amabilis. The findings were similar to those of previous studies on R. amabilis, where R. amabilis formed a small branch with R. takesimensis and R. crataegifolius, all of which are members of the Rosoideae (Yang et al. Citation2020). The complete chloroplast data of R. irritans could be used as a reference for taxonomic studies on this genus. The cp genome information could also be used in phylogeny, DNA barcoding, and conservation genetics.
Author contributions
Yong-Xia Han, Li Tong and Zong-Hao Zhang accomplished the design, writing, and revision of this study. Shi-Bing Yang and Fang Yang participated in the collection and identification of plant material. All authors read and approved the final manuscript and agreed to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under the accession no. MN652919. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA771538, SRR16529182, and SAMN22315382, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Cho MS, Kim Y, Kim SC, Park J. 2019. The complete chloroplast genome of Korean Pyrus ussuriensis Maxim. (Rosaceae): providing genetic background of two types of P. ussuriensis. Mitochondrial DNA B Resour. 4(2):2424–2425.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Editorial Committee of Flora of China, Chinese Academy of Sciences. 1985. Flora of China. Vol. 37. Beijing, China: Science Press.
- Kim Y, Heo KI, Nam S, Xi H, Lee S, Park J. 2019. The complete chloroplast genome of candidate new species from Rosa rugosa in Korea (Rosaceae). Mitochondrial DNA B Resour. 4(2):2433–2435.
- Kim HW, Kim KJ. 2016. The complete plastome sequence of Pentactina rupicola Nakai (Rosaceae), a genus endemic to Korea. Mitochondrial DNA B Resour. 1(1):698–700.
- Li WL, He SA, Gu Y. 2000. An outline on utilization value of Chinese bramble (Rubus L.). Journal of Wuhan Botanical Research. 18(3):237–243.
- Li Y, Liu Y, Wu P, Zhou S, Wang L, Zhou S. 2020. The complete chloroplast genome sequence of Malus toringoides (Rosaceae). Mitochondrial DNA B Resour. 5(3):2787–2789.
- Lim SY, Jang JH, Lee HJ, Park SS, Kim SR, Lee KM, Kim JK, Park H, Jung HK. 2021. Characteristics and phylogenetic analysis of the complete chloroplast genome of Paeonia japonica (Paeoniaceae). Mitochondrial DNA B Resour. 6(3):734–735.
- Liu XJ, Wang XR, Tang HR, Chen Q. 2021. The complete chloroplast genome sequence of a hybrid blackberry (Rubus spp.) cultivar. Mitochondrial DNA B Resour. 6(8):2103–2104.
- Liu Y, Xiang S, Fu X. 2021. Characterization of the complete chloroplast genome sequence of medicinal plant: potentilla bifurca (Rosaceae). Mitochondrial DNA B Resour. 6(1):143–144.
- Park J, Heo KI, Kim Y, Kwon W. 2019. The complete chloroplast genome of Potentilla Freyniana Bornm. (Rosaceae). Mitochondrial DNA B Resour. 4(2):2420–2421.
- Shi LC, Chen HM, Jiang M, Wang LQ, Wu X, Huang LF, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 57(5):758–771.
- Wang X, Wang J, Luo M. 2020. Characterization of the complete chloroplast genome of Prunus davidiana, an excellent horticultural species. Mitochondrial DNA B Resour. 5(1):932–933.
- Yang F, Zhang ZH, Tong L. 2020. The complete chloroplast genome sequence of Rubus amabilis Focke. Mitochondrial DNA B Resour. 5(2):1975–1976.
- Zhang G, Liu Y, Hai P. 2021. The complete chloroplast genome of Tibetan medicinal plant Rubus phoenicolasius Maxim. Mitochondrial DNA B Resour. 6(3):886–887.
- Zhang PP, Wang L, Lu X. 2022. Complete chloroplast genome of Geum aleppicum (Rosaceae). Mitochondrial DNA B Resour. 7(1):234–235.
