Abstract
In this study, we sequenced, annotated, and characterized the mitogenome of Tanichthys albiventris for the first time. The complete mitogenome is 16,544 bp in length, consisting of 13 protein-coding genes (PCGs), 22 transfer RNA genes, and two ribosomal RNA genes. We found nine genetic overlaps and 17 intergenic spacer regions throughout the mitogenome of T. albiventris. The A + T content of the mitogenome is 60.93%. All PCGs have the same start codon of the standard ATG, excepting for that of cytochrome c oxidase 1 (cox1) which is the GTG. A phylogenetic analysis with another 15 species of the Cyprinidae was performed using MrBayes and IQtree, based on the amino acid sequences of 13 mitochondrial PCGs. The results indicated that T. albiventris shares a close ancestry with Tanichthys albonubes.
Tanichthys is a small freshwater fish belonging to the family Tanichthyidae within the superfamily Cyprinoidea (Chen and Mayden Citation2009; Mayden and Chen Citation2010; Stout et al. Citation2016). It has become a famous ornamental fish owing to its colorful appearance (Weitzman and Chan Citation1966). Tanichthys are distributed in several basins in southeastern China, and northern and central Vietnam (Lin Citation1932; Freyhof and Herder Citation2001; Thang et al. Citation2019; Li et al. Citation2022). The wild populations of Tanichthys have been classified as a second-class protected animal in China due to habitat loss (Yue and Chen Citation1998). To date, 12 valid species have been reported within the genus Tanichthys (Freyhof and Herder Citation2001; Thang et al. Citation2019; Jin et al. Citation2022; Li et al. Citation2022). Tanichthys albiventris Li, Bohlen & Liao, 2022 can be distinguished from all other congeners by the number of branched dorsal- and anal-fin rays, and the color of dorsal- and anal-fin margins (Li et al. Citation2022).
In this study, the complete mitochondrial genome of T. albiventris was sequenced and characterized. The samples of T. albiventris () were collected from the Jiangping River (E108.0811 and N21.6548) in Jiangping Town, Dongxing City, Guangxi Province, China, and stored in Xinyang Agricultural and Forestry University Aquatic Museum (https://www.xyafu.edu.cn/, Liangjie Zhao and [email protected]), voucher number XYAFU-Mu-1804158. Prior to further processing, samples were placed in 100% ethanol during collection and stored at −80 °C before DNA extraction. Total DNA was extracted from caudal fin tissue using the Ezup Column Animal Genomic DNA Purification Kit (Sangon Biotech (Shanghai) Co., Ltd., Shanghai, China). The quality of the isolated DNA was detected by 1.2% agarose gel electrophoresis and the DNA was stored at −20 °C until utilization. Long and accurate PCR (LA PCR) was performed using a set of specific primers to amplify the complete mitochondrial genome sequence. Based on the sequenced and published sequences of other Tanichthys species in the NCBI database, nine pairs of specific primers were successfully screened in this study, and the amplified products ranged from 864 bp to 3829 bp, covering the entire length of the mitochondrial gene sequence. PCR products were sequenced at Shanghai Biotech using an ABI 3730XL DNA analyzer (Applied Biosystems Inc., Foster City, CA).
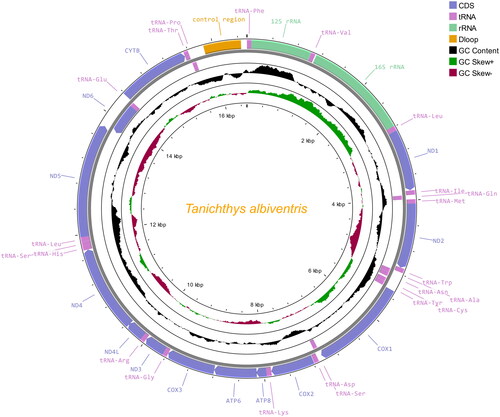
The complete mitochondrial genome of T. albiventris is 16,544 bp in length and deposited in GenBank with the accession no. OM687295. It consists of 13 protein-coding genes (PCGs), 22 tRNA genes, and two rRNA genes (). Twelve of the PCGs are located in the heavy strand, while only the gene encoding nad6 is located in the light strand. Similar to the preference of PCGs located in the heavy strand, 14 of the 22 tRNA genes are also located in the heavy strand. The ATG is commonly used as the start codon in this mitogenome with the exception of cytochrome c oxidase 1 (cox1) which starts with the GTG. Except for the incomplete stop codon of ‘T–’ in cox2 and cob as well as that of ‘TA–’ in cox3 and nad4, the remaining PCGs all stop with the TAG or TAA. The 22 tRNA genes were found to fold into a typical cloverleaf secondary structure by the analysis with the MITOS web server (Bernt et al. Citation2013). The overall basic nucleotide composition of the heavy strand in T. albiventris consists of 31.62% A, 29.31% T, 15.34% G, and 23.73% C, with an A + T content of 60.93%.
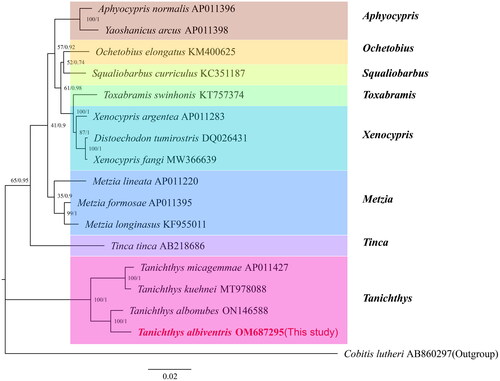
A total of 16 species (including this study species) from eight genera of Cyprinidae fishes were selected, and the protein sequences of the 13 PCGs were used to construct a phylogenetic evolutionary relationship tree. The mitogenome of Cobitis lutheri (accession no. AB860297) was chosen as the outgroup data (Maddison et al. Citation1984). The phylogenetic analysis was performed by MrBayes v3.2.6 and IQtree v1.6.10 (Ronquist et al. Citation2012; Minh et al. Citation2020). PartitionFinder 2.1.1 was employed to select the optimal evolutionary models for phylogenetic analysis (Lanfear et al. Citation2017). The resulting phylogenetic trees can be directly viewed in FigTree v1.4.2.
The constructed phylogenetic tree based on the optimal model is shown in . The results indicated that T. albiventris is most closely related to T. albonubes, and they both clustered with Tanichthys micagemmae and Tanichthys kuehnei. The molecular phylogenetic analysis of the present study support the attribution of T. albiventris to the genus Tanichthys (Li et al. Citation2022), and the monophyly of the family Tanichthyidae (Mayden and Chen Citation2010; Stout et al. Citation2016), but disagree with the result of Tang et al. (Citation2010) that the genera Tanichthys and Tinca constitute a monophyletic family. The comparative mitogenomic analysis of T. albiventris may provide valuable phylogenetic information.
Figure 3. Phylogenetic tree constructed using the IQtree and MrBayes method based on the amino acid sequences of 13 proteins translated by the mitochondrial genome. The numbers on the branches indicate the posterior probabilities (IQtree/MrBayes). The red font represents the target species in this study.
Ethical approval
Experiments were performed in accordance with the recommendations of the Ethics Committee of Xinyang Agriculture and Forestry University. These policies were enacted according to the Chinese Association for the Laboratory Animal Sciences and the Institutional Animal Care and Use Committee (IACUC) protocols.
Author contributions
Conceived and designed the experiments: Fan Li and Liangjie Zhao. Performed the experiments: Tiezhu Yang, Dongpu Li, and Wenhao Sun. Sample collection: Fan Li, Liangjie Zhao, Wenhao Sun, and Dongpu Li. Data acquisition: Chenxi Tan, Dongpu Li, and Wenhao Sun. Analyzed the data: Tiezhu Yang, Chenxi Tan, and Liangjie Zhao. Wrote the manuscript: Tiezhu Yang, Liangjie Zhao, and Fan Li. Helped to proofread the manuscript: Chenxi Tan, Liangjie Zhao, and Fan Li. All authors have read and agreed to the published version of the manuscript.
Acknowledgements
We appreciate anonymous reviewers for providing valuable comments on this article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. OM687295. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA887552, SRR21820798, and SAMN31172242, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chen WJ, Mayden RL. 2009. Molecular systematics of the Cyprinoidea (Teleostei: Cypriniformes), the world’s largest clade of freshwater fishes: further evidence from six nuclear genes. Mol Phylogenet Evol. 52(2):544–549.
- Freyhof J, Herder F. 2001. Tanichthys micagemmae, a new miniature cyprinid fish from central Vietnam (Cypriniformes: Cyprinidae). Ichthyol Explor Freshw. 12:215–220.
- Jin J, Li C, Zhao J. 2022. Descriptions of six new species in White Cloud Mountain minnow Tanichthys albonubes complex (Cypriniformes: Tanichthyidae) in southern China. J Fish Biol. 100(4):1062–1087.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773.
- Li F, Liao T-Y, Bohlen J, Shen Z-X, Zhao L-J, Li S. 2022. Two new species of Tanichthys (Teleostei: Cypriniformes) from China. J Verteb Biol. 71(21067):1–13.
- Lin S. 1932. New cyprinid fishes from white cloud mountain, Canton. Lingnan Sci J. 11:379–383.
- Maddison WP, Donoghue MJ, Maddison DR. 1984. Outgroup analysis and parsimony. Syst Biol. 33(1):83–103.
- Mayden RL, Chen WJ. 2010. The world’s smallest vertebrate species of the genus Paedocypris: a new family of freshwater fishes and the sister group to the world’s most diverse clade of freshwater fishes (Teleostei: Cypriniformes). Mol Phylogenet Evol. 57(1):152–175.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Stout CC, Tan M, Lemmon AR, Lemmon EM, Armbruster JW. 2016. Resolving Cypriniformes relationships using an anchored enrichment approach. BMC Evol Biol. 16(1):244.
- Tang KL, Agnew MK, Hirt MV, Sado T, Schneider LM, Freyhof J, Sulaiman Z, Swartz E, Vidthayanon C, Miya M, et al. 2010. Systematics of the subfamily Danioninae (Teleostei: Cypriniformes: Cyprinidae). Mol Phylogenet Evol. 57(1):189–214.
- Thang HN, Bohlen J, Dvorák T, Šlechtová V. 2019. Tanichthys kuehnei, new species, from Central Vietnam (Cypriniformes: Cyprinidae). Ichthyol Explor Freshw. 1081:1–10.
- Weitzman SH, Chan LL. 1966. Identification and relationships of Tanichthys albonubes and Aphyocypris pooni, two cyprinid fishes from South China and Hong Kong. Copeia. 1966(2):285–296.
- Yue PQ, Chen YY. 1998. China red data book of endangered animals: Pisces. Beijing: Science Press.