Abstract
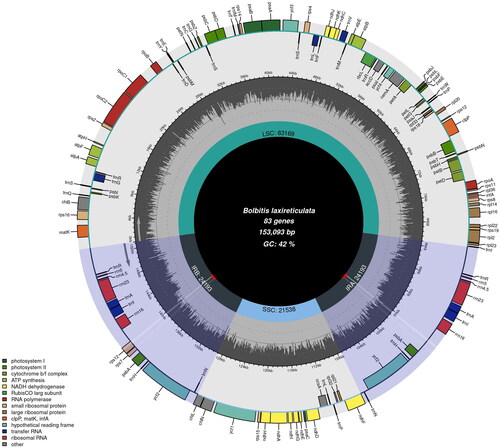
Bolbitis laxireticulata is a potential ornamental plant, which is restricted to eastern Asia. Here, we sequenced the complete chloroplast (cp) genome of B. laxireticulata and constructed a phylogenetic cp tree of Dryopteridaceae to study their relationships. The cp genome of B. laxireticulata is 153,093 bp in length, being made up of large single-copy (LSC, 83,169 bp), small single-copy (SSC, 21,538 bp), and a pair of region inverted repeats (IRs, 24,193 bp). It has 124 genes including 83 protein-coding genes, 33 tRNA genes, and eight rRNA genes. With the maximum-likelihood tree indicating, B. laxireticulata is more closely related to B. subcordata.
The genus Bolbitis Schott (Dryopteridaceae) is distributed in tropical and subtropical regions which includes around 80 species (Schuettpelz Citation2016). With viviparous apices of fronds and variable shape that resembles both Bolbitis sinensis_ (Baker) K. Iwatsuki Citation1959 and B. singaporensis_Holttum 1928, B. laxireticulata_ K. Iwatsuki Citation1959 has higher ornamental value, and restricted to Guangdong, Hainan, Taiwan (China) and Ryukyu Islands (Japan) (Iwatsuki Citation1959; Dong and Zhang Citation2005). It is the first time to sequence the cpDNA of B. laxireticulata. Here, we constructed a chloroplast (cp) maximum-likelihood (ML) tree and aimed to explore its genetic relationships with other species in Bolbitis. Research on the relationship of those species can not only provide necessary evidence for combing some complex system evolutionary relationships, but also can solve hybrid transition form and the classification of the difficult problems that cause. This study provides essential basic data for the formation and reticulate evolution of the Bolbitis, and provides theoretical basis and technical support for the further work of species conservation and breeding.
The fresh leaf materials of B. laxireticulata were collected in Longmen County (longitude: 113°28′52″E, latitude: 23°54′09″N, altitude: 550 m), Guangdong Province, China (). The voucher specimen was deposited at Herbarium of South China Botanical Garden, Chinese Academy of Sciences (IBSC) (http://herbarium.scbg.cas.cn/, Feiyan Zeng, [email protected]), under the voucher number: WFG6462. CTAB methods were used to extract the total genomic DNA from the leaf tissue samples (Doyle Citation1987). Preparation and sequencing of genomic libraries used the Illumina NovaSeq 6000 platform (Illumina Inc., San Diego, CA) with 150 bp end lengths. The resultant sequences were filtered and the raw data were removed by Trimmomatic (Bolger Citation2014). The adaptor-free reads were then assembled with SPAdes v3.11 (Bankevich et al. Citation2012). GeSeq v2.03 (Michael Citation2017) was used to annotate the cp genome. Then, the genome was deposited in GenBank (accession number: OM350395).
Figure 1. Species reference images of Bolbitis laxireticulata. (A) Plant shape of B. laxireticulata. (B) Morphological characteristics of leaves of B. laxireticulata (lobes varied in length and irregular, reticulate veins conspicuous). The species photo was taken by the author in Nankunshan Nature Reserve, Longmen Country, Huizhou, China, August 2021, without any copyright issues.

The raw reads of B. laxireticulata are approximately 6 G, and its cp genome is 153,093 bp in length, including a large single-copy (LSC, 83,169 bp), a small single-copy (SSC, 21,538 bp), and a pair of inverted repeats (IRs, 24,193 bp). Totally, 124 genes were annotated, including 83 protein-coding genes, 33 tRNA genes, and eight rRNA genes, within which 14 genes (atpF, ndhA, petB, petD, rpl2, rpl16, rpoC1, rps12, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnL-UAA, trnA-UAC) had one intron, two genes (clpP, ycf3) had two introns (). The overall G/C content of total genome is 42.3%.
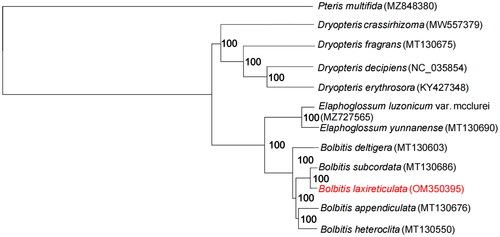
In order to understand the phylogenetic relationship of B. laxireticulata, we downloaded 11 cp genomes from GenBank to build phylogenetic relationship in a phylogram (). Pteris multifida_ Poir 1804 was chosen as the outgroup. Geneious prime and Mauve plugin were used to adjust gene order and remove a copy of the IR regions of the cp genome to detect gene rearrangements with homozygous genes. We used MAFFT v.7 (Katoh and Standley Citation2013) to align the 12 cp genes which is used to construct the phylogenetic tree. Then we used TrimAl (Capella-Gutierrez et al. Citation2009) to trim the alignment with automatic parameter. Finally, IQ-TREE v.1.4.2 (Nguyen et al. Citation2015) program was used to run an ML tree with 5000 bootstrap replicates (). All GenBank accession numbers including the new obtained accession number of B. laxireticulata are given in . We then generated a physical map of the cp genome using Chloroplast Genome Viewer (CPGView) (http://www.1kmpg.cn/cpgview). The ML tree shows that B. laxireticulata is more closely related to B. subcordata_ (Cop.) Ching 1934 (), and also has closer phylogenetic relationship to B. appendiculata_ (Willdenow) K. Iwatsuki Citation1959 and B. heteroclita (Presl) Ching 1934. The next step will be to further verify the hybridization relationship of B. laxireticulata at macro- and micro-levels.
Author contributions
Fa-Guo Wang, Yu-Jie Liao, and Li-Yun Nie involved in the conception and design or analysis and interpretation of the data. Yu-Jie Liao and Fa-Guo Wang drafted and modified paper. Wen-Chao Zhong, Ai-Hua Wang, Xin-Xin Cheng, and Lei Duan were involved in design of the work, assisting with sampling and paper edition, some suggestions and opinions were given by them. All the authors agreed to be responsible for all aspects of the work.
Ethics statement
This study includes no human, animal, or endangered plant samples, and the sample was legally collected in accordance with guidelines provided by the authors’ institution and national or international regulations. Field studies complied with local legislation. No ethical approval/permission is required for this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. OM350395. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA817290, SRR18361020, and SAMN26754306, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Capella-Gutierrez S, Silla-Martınez JM, Gabaldon T. 2009. TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Dong SY, Zhang XC. 2005. A taxonomic revision of the fern genus Bolbitis (Bolbitidaceae) from China. Acta Phytotaxon Sin. 43:97–115.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Iwatsuki K. 1959. Taxonomic studies of Pteridophyta III. Acta Phytotaxon Geobot. 18(2–3):44–59.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Michael T, Pascal L, Tommaso P, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Schuettpelz E, Schneider H, Smith AR, Hovenkamp P, Prado J, Rouhan G, Salino A, Sundue M, Almeida TE, Parris BS, et al. 2016. A community-derived classification for extant lycophytes and ferns. J Syst Evol. 54(6):563–603.