Abstract
Senna alata, a flowering shrub, is widely cultivated in the Philippines for its anti-fungal properties. Despite this, its chloroplast genome is not yet established. We assembled and annotated the complete chloroplast genome of accession from the germplasm collection of the Institute of Crop Science, University of the Philippines, Los Baños, using Illumina sequencing data. The complete cp genome was 159,176-bp long characterized by a large single copy of 88,769 bp, short single-copy of 18,301 bp and a pair of inverted repeat regions of 26,053 bp each. The overall GC content of the chloroplast genome was 36.4%. The plastome comprised 37 tRNA genes, 8 rRNA genes and 78 mRNA genes. Phylogenetic analysis showed that S. alata is closely related to S. siamea.
Introduction
Senna alata (Linnaeus) Roxburgh 1832, also known as candle bush, is a flowering ornamental shrub under subfamily Caesalpinioideae of family Fabaceae (Leguminosae). The species is native to Colombia, Venezuela, the Guyanas and Brazil (Irwin and Barneby Citation1982) and also commonly found in Asia and Africa (Kumar et al. Citation2008). It was introduced in the Philippines during the Spanish colonial period, ca. 16th–eighteenth century, via the Galleon trade (Amano et al. Citation2021). S. alata is locally called “akapulko” in the country, and it is widely dispersed and cultivated for medicinal purposes (Palanichamy and Nagarajan Citation1990; Philippine Pharmacopeia 1 [PPI], Citation2004). It has been used to treat diseases due to its antifungal (Oladeji et al. Citation2016), laxative (Adelowo and Oladeji Citation2017), anti-helmintic (Kundu et al. Citation2012) and anti-inflammatory (Sagnia et al. Citation2014) properties. Akapulko is included in the Department of Health (DOH)-Philippines list of ten recommended medicinal plants that had been validated for their safety and efficacy (Zarsuelo et al. Citation2018). Despite these promising health-promoting benefits, no investigation has characterized the complete chloroplast genome sequence; hence, in this study, we assembled and annotated the complete chloroplast of S. alata, which will be useful in elucidating evolutionary and phylogenetic relationships under the subfamily Caesalpinioideae of the Fabaceae family.
Materials and methods
We collected fresh leaves of S. alata from the field genebank of the Crop Breeding and Genetic Resources Division, Institute of Crop Science, University of the Philippines Los Baños (UPLB), Laguna, Philippines with a type locality at Nueva Ecija, Philippines (15° 22′ 04.0” N 121° 03′ 26.0” E). We characterized the morphology of the accession (ICROPS 2019 149) based on the identified distinguished morphological markers in the Senna group (Roxburgh and Wallich Citation1820) for future taxonomic revisions. This is also to ensure that the genotype is associated with a particular reference specimen conserved in the genebank and preserved in the herbarium. The whole plant, leaves, inflorescence and pods of the accession in the field genebank were also photographed using the Nikon COOLPIX S9500 Digital Camera. The quantitative measurements reported in this paper are computed average values. The voucher specimen (ICROPS 2019 149) of this accession was deposited in the Philippine Herbarium of Cultivated Plants, UPLB (https://cafs.uplb.edu.ph/icrops/, Renerio P. Gentallan Jr., [email protected]). The genomic DNA from fresh samples was extracted using the slightly-modified CTAB method (Doyle and Doyle Citation1987) and sent to NovogeneAIT Genomics Singapore PTE LTD, Singapore, for sequencing using the HiSeq-PE150 platform (Illumina Inc., San Diego, CA, USA). The 150-bp pair-end raw reads were produced which were successively filtered to generate 18,652,000 of cleaned reads. We used the GetOrganelle v1.7.5+ software (Jin et al. Citation2020) to assemble the chloroplast genome which generated a circular genome. Subsequently, this genome was annotated and mapped using CPGAVAS2 (Shi et al. Citation2019) and was visualized using Chloroplast Genome Viewer (CPGView) (Liu et al. Citation2023). The assembled chloroplast genome sequence was submitted to the GenBank (accession no. ON653612/NC_065665.1) of the National Center for Biotechnology Information (NCBI).
Results and discussion
Morphological characterization of the plant material
S. alatais 2.29 m tall with a 1.55 m spread. Leaflet () opposite, 8 pairs, blade obovate, retuse apex, dark green with a yellow midrib and orange margin color, 9.82 × 4.6 cm. Petal () ovate, 15.02 cm long while bract, pedicel and rachis () are 12.48, 6.34 and 37.3 cm long, respectively. Pod () luster matte with a strong degree of ridge depth, 11.82 × 1.76 cm. Seed () surface wrinkled, glossy, black, 11.83 × 4.41 × 2.09 mm. The characteristics of the accession (ICROPS 2019-149) used for the assembly fall within the range of the description established for S. alata (Roxburgh and Wallich Citation1820). Hou et al. (Citation1996) reported that S. alata is differentiated from other species through its characteristic pedicel that is shorter than the sepal and pods (), which are winged.
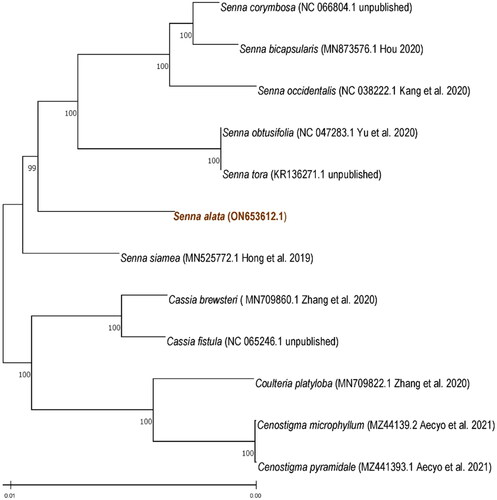
Complete chloroplast genome and phylogenetic anlaysis of S. alata
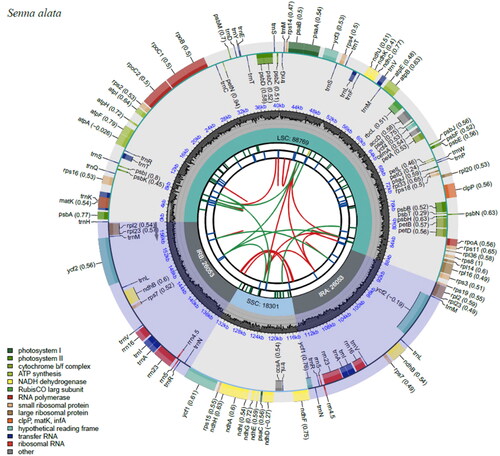
The chloroplast genome of S. alata had a sequence length of 159,176 bp which exhibited a typical quadripartite circular structure (, Figure S1). The sequence included a large single copy (LSC), a short single-copy (SSC) and two inverted repeat regions (IRa and IRb) corresponding to 88,769, 18,301 and 26,053 bp, respectively. The overall GC content of the chloroplast genome was 36.4% with base compositions of 31.4% A, 32.2% T, 17.9% G and 18.5% C. S. alata had a shorter sequence length compared to S. bicapsularis with 161,056 bp (Hou Citation2020), S. spectabilis with 162,754 bp (Shi et al. Citation2020), S. tora with 161,050 bp (Xu et al. Citation2020), S. occidentalis with 159,993 bp (Kang et al. Citation2020) and S. obtusifolia with 162,426 bp (Yu et al. Citation2020). The chloroplast genome of S. alata comprised of 123 genes coding for 37 tRNAs, 8 rRNAs and 78 mRNAs. Among these are 42 genes for photosynthesis, 27 genes for self-replication, three conserved ORF (ycf1, ycf1, ycf4), and 6 other genes (accD, ccsA, cemA, clpP, infA and matK). In addition, 13 cis-splicing genes were observed, out of which 9 were unique (rps16, atpF, rpoC1, ycf3, clpP, petB, petD, rpl16, ndhA) and 2 were duplicated (rpl2 and ndhB) (Figure S1).
Figure 2. Chloroplast genome map of S. alata. The map contains seven circles, where the first circle (from the center going outward) shows the distributed repeats connected with red (the forward direction) and green (the reverse direction) arcs. The second circle shows the tandem repeats marked with short bars. The next circle shows the microsatellite sequences as short bars. The fourth circle shows the size of the LSC and SSC. The fifth circle shows the IRA and IRB while the sixth circle shows the GC contents along the plastome. The seventh circle shows the genes having different colors based on their functional groups.
The coding sequences from the chloroplast genomes of S. alata and twelve other related species (NC 047283.1, KR136271.1, MN873576.1, NC 038222.1, MN525772.1, MN709860.1, MN709822.1, MZ441394.1, MZ441392.1, MZ441393.1, NC_065246.1, NC_066804.1) were downloaded from the NCBI database to construct a phylogenetic tree. These were aligned using MAFFT (Katoh and Standley Citation2013) and phylogenetic analysis was performed using MEGA-X (Kumar et al. Citation2018). These methods generated a Maximum Likelihood (ML) tree with the General Time Reversible, Gamma-Invariant (GTR + G + I) as the model (Nei and Kumar, Citation2000) with 1,000 bootstrap replicates. The phylogenetic analysis showed two major clades, the Senna clade and the outgroups including Cassia and Cenostigma species (). The phylogenetic tree also shows a close relationship between S. alata and S. siamea.
Conclusion
The complete chloroplast genome of S. alata was sequenced and the assembled genome has a sequence length of 159,176 bp which was shorter than the other Senna species. Phylogenetic analysis showed a close relationship between S. alata and S. siamea. The data will contribute to the elucidation of phylogenetic relationships under the genus Senna of the Fabaceae family.
Author contributions
All authors agree to be accountable for all aspects of the work. Kristine Joyce Quiñones and Renerio Gentallan Jr. conceptualized the study, performed the experiments, analyzed the data, and wrote the paper; Michael Cedric Bartolome, Roselle Madayag, Juan Rodrigo Vera Cruz, Jessabel Magtoltol, Reneliza Cejalvo, Bartimeus Buiene Alvaran, Angeleigh Rose Cirunay and Emmanuel Bonifacio Timog planted, collected, photographed and prepared the germplasm, pressed the herbarium samples, reviewed literature, performed experiments, edited the paper; Teresita Borromeo, Nestor Altoveros, and Leah Endonela, helped conceptualized the study, validated the design of the experiment and the data presented, reviewed drafts of the paper.
Ethical approval
The collection of plant material was carried out in accordance with guide-lines provided by the authors’ institution (Institute of Crop Science, College of Agriculture and Food Science, University of the Philippines Los Baños).
Supplemental Material
Download MS Word (278.1 KB)Acknowledgments
The study would like to thank the Department of Science and Technology- Philippine Council for Agriculture, Aquatic and Natural Resources Research and Development (DOST-PCAARRD) for the support. The authors would also like to thank Ronil Beliber and Arvin Medrano for cultivating the germplasm, and Eddelaine Joyce Bautista and Edna Mercado for processing the paperwork needed for the research project.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.govunder the accession no. ON653612. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA867486, SRR20980268 and SAMN30201905, respectively.
Additional information
Funding
References
- Adelowo F, Oladeji O. 2017. An overview of the phytochemical analysis of bioactive compounds in S. alata. American Chemical and Biochemical Engineering. 2(1):7–14.
- Aecyo P, Marques A, Huettel B, Silva A, Esposito T, Ribeiro E, Leal IR, Gagnon E, Souza G, Pedrosa-Harand A. 2021. Plastome evolution in the Caesalpinia group (Leguminosae) and its application in phylogenomics and populations genetics. Planta. 254(2):1–9.
- Amano N, Bankoff G, Findley DM, Barretto-Tesoro G, Roberts P. 2021. Archaeological and historical insights into the ecological impacts of pre-colonial and colonial introductions into the Philippine Archipelago. The Holocene. 31(2):313–330.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Hou D, Larsen K, Larsen SS. 1996. Caesalpiniaceae (Leguminosae-Caesalpinioideae). Flora Malesiana-Series 1, Spermatophyta 1. 12(2):409–730.
- Hou Z. 2020. The complete chloroplast genome sequence of Senna bicapsularis. Mitochondrial DNA B. 5(3):2095–2096.
- Irwin HS, Barneby RC. 1982. The American Cassiinae: a synoptical revision of Leguminosae tribe Cassieae subtribe Cassiinae in the New World. New York Botanical Garden. v +:918. pp.
- Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de nozvo assembly of organelle genomes. Genome Biol. 21(1):1–31.
- Kang S, Lee H, Kim C, Chang S, Kang JM, Lee SM. 2020. The complete chloroplast genomes of the medicinal plants, Senna tora and Senna occidentalis species. Mitochondrial DNA Part B. 5(2):1673–1674.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar A, Shukla R, Singh P, Prasad CS, Prasad NK. 2008. Assessment of Thymus vulgaris L. essential oil as a safe botanical preservative against post-harvest fungal infestation of food commodities. Innov Food Sci Emerg Technol.. 9(4):75–580.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Kundu S, Roy S, Lyndem LM. 2012. Cassia alata L: potential role as anthelmintic agent against Hymenolepis diminuta. Parasitol Res. 111(3):1187–1192.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 0:1–11.
- Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. USA (NY): Oxford University Press,
- Oladeji SO, Adelowo FE, Odelade KA. 2016. Mass spectroscopic and phytochemical screening of phenolic compounds in the leaf extract of Senna alata (L.) Roxb. Evidence-based complementary and alternative medicine 9 (Fabales: fabaceae). Braz J Biol Sci.. 3(5):209–219.
- Palanichamy S, Nagarajan S. 1990. Analgesic activity of Cassia alata leaf extract and kaempferol 3-o-sophoroside. J Ethnopharmacol. 29(1):73–78.
- Philippine Pharmacopeia 1 [PPI] 2004. Philippine Pharmacopeia, 1. Bureau of Food and Drugs, Department of Health and Japan International Cooperation Agency. Manila: Himiko Arts and Concepts
- Roxburgh W, Wallich N. 1820. Flora indica: or, descriptions of Indian plants. Serampore, India: Printed at the Mission Press.
- Sagnia B, Fedeli D, Casetti R, Montesano C, Falcioni G, Colizzi V. 2014. Antioxidant and anti-inflammatory activities of extracts from Cassia alata, Eleusine indica, Eremomastax speciosa, Carica papaya and Polyscias fulva medicinal plants collected in Cameroon. PLoS One. 9(8):e103999.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Shi Z, Shi G, Zhao K, Sun B. 2020. Complete chloroplast genome of Senna spectabilis (DC.) H.S. Irwin & Barneby (Fabaceae) and phylogenetic analysis. Mitochondrial DNA B Resour. 5(3):2846–2847.
- Xu Q, Ma L, Chen G. 2020. The complete chloroplast genome sequence of Senna tora and phylogenetic analysis. Mitochondrial DNA Part B. 5(3):3433–3435.
- Yu X, Tan W, Gao H, Miao L, Tian X. 2020. Development of a specific mini-barcode from plastome and its application for qualitative and quantitative identification of processed herbal products using DNA metabarcoding technique: a case study on Senna. Front Pharmacol. 11:585687.
- Zarsuelo MAM, Zordilla ZD, Anacio DB. 2018. Review of regulatory policies on and benefits of herbal medicine in the Philippines. Acta Med Philippina. 52:473–479.