Abstract
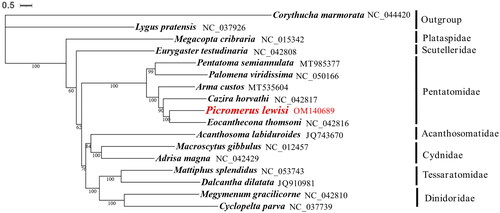
Picromerus lewisi Scott (Hemiptera: Pentatomidae) is a widely used natural enemy, through this study, we proved that its complete mitochondrial genome of it had similar characteristics to those of other Hemiptera. The mitogenome of P. lewisi is a circular molecule of 18,123 bp with 74.0% A + T content, containing 13 protein-coding genes (PCGs), 22 tRNAs, 2 rRNAs, and one control region. Phylogenetic tree based on 13 PCGs from 17 Panheteroptera species (two species of the Cimicomorpha are used as outgroup, 15 species belong to the Pentatomomorpha) suggested that P. lewisi has a closer relationship with E. thomsoni within Pentatomidae family.
Picromerus lewisi Scott 1874 is an important generalist pentatomid predator of multiple species of pests, especially for lepidopteran pests (Tang et al. Citation2018; Wang et al. Citation2019, Citation2020; Fu et al. Citation2021; Shen et al. Citation2022), and has been used in biological control programs with its large predatory capacity, strong activity and adaptability in daily living, and suitable for artificial rearing (Yang et al. Citation2018). In this study, we sequenced the complete mitochondrial genome of Picromerus lewisi Scott. Surviving adult individuals were fed in the artificial climate chamber of Kaiyang country, Guiyang, Guizhou province of China (106°57′14′′E, 27°16′34′′N) (). Presently, the sample was deposited in the Institute of Entomology, Guizhou University, Guiyang, China (voucher Gzh-2021-026, Hai-yan Zhao, [email protected]). We used the DNeasy Blood and Tissue Kit (cat.nos.69504 and 69506, QIAGEN, Hilden, Germany) to extract the total DNA from the head of the adult. An Illumina ReSeq library was produced, with an average insert size of 300 bp (a paired-end length of 150 bp). The library was then sequenced using the Illumina Novaseq6000 platform (Berry Genomics, Beijing, China). We assembled and annotated the complete mitogenome sequence using NOVOPlasty v2.7.2 (Dierckxsens et al. Citation2017), with the default setting for the K-mer value, and MitoZ v2.4-alpha (Meng et al. Citation2019) was used to annotate the genome.
The mitochondrial genome of P. lewisi was 18,123 bp long (GenBank accession OM140689) and consisted of 13 PCGs, 22 tRNA genes, two rRNA genes (lrRNA and srRNA), and one control region. The length of lrRNA and srRNA is 1,276 bp and 800 bp in length. The lengths of the 22 tRNA genes ranged from 63 bp (trnC) to 74 bp (trnK) ().
Figure 2. Genome map of P. lewisi. Mitochondrial genome map was drawn using the Chloroplot (https://irscope.shinyapps.io/Chloroplot/).
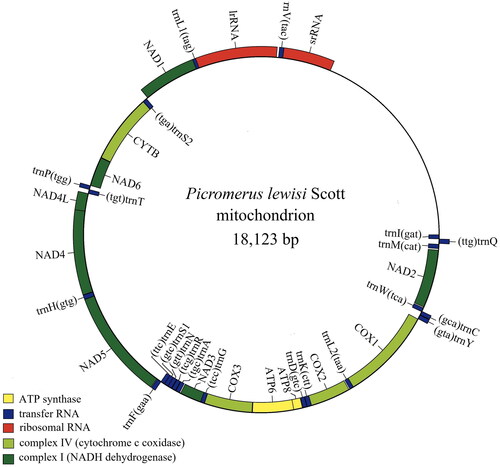
The overall base composition of the mitogenome was estimated as follows: A 40.9%, T 33.1%, C 15.3%, and 10.7 8.0%, with a high A + T content of 74.0%. Among the 13 PCGs, twelve PCGs started with a typical ATN codon: five with ATT (NAD2, NAD4L, NAD5, NAD6, COX2), four with ATG(ATP6, COX3, NAD4, CYTB), three with ATA(ATP8, NAD1, NAD3), whereas COX1 started with TTG. Most of the PCGs terminated with the stop codon TAA, CYTB ended with stop codon TAG, whilst COX2 uses a single T residue as an incomplete stop codon. The A + T content of 22 tRNA of Picromerus lewisi Scott ranged from 60.6% (trnM) to 89.1% (trnL2). The A + T content of IrRNA and srRNA are 76.6% and 76.1%, respectively.
Based on the concatenated amino acid sequences of 13 PCGs from 17 Panheteroptera species, PhyloSuite v1.2.2 (Zhang et al. Citation2020) was used for phylogenetic analysis, a phylogenetic tree was constructed using Bayesian inference (BI) in MrBayes (Huelsenbeck and Ronquist Citation2001). The iTOL webtool (https://itol.embl.de) (Letunic and Bork Citation2007) was used to view the phylogenetic tree. Corythucha marmorata and Lygus pratensis were used as outgroups. Our analysis positioned P. lewisi in a well-supported clade with Eocanthecona thomsoni. It was shown that P. lewisi has a closer relationship with E. thomsoni within Pentatomidae family, which accord with the traditional taxonomy. In conclusion, the complete mitogenome of P. lewisi is decoded in this study and provides essential and important DNA molecular data for further phylogenetic and evolutionary analysis for Pentatomidae ().
Ethical approval
This study does not need ethical approval or permission to collect the sample.
Authors contributions
Xiao-han Chen completed the drafting of the manuscript. Meng Leng analyzed the data. Mao-fa Yang and Hai-yan Zhao designed and revised it critically for intellectual content. Di-hui Yao collected the adult and carried out the study.
Acknowledgments
The authors are thankful to acknowledge the comments of the anonymous reviewers.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under accession no. OM140689. The associated BioProject, SRA, and BioSample numbers are PRJNA890215, SRR21891108, and SAMN31269902, respectively.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Fu CY, Xu TM, Wen SH, Zhang SA, Liu F, Du GZ, Chen B, Zhang LM. 2021. Predation behaviors and functional responses of Picromerus lewisi to the Larvae of Ostrinia furnacalis. Chin J Biol Control. 37(5):956–962.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755.
- Letunic I, Bork P. 2007. Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 23:127–128.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63–e63.
- Shen XX, Tian TA, Liu JF, Yu XF, Dong XL, Li ZM, Yang MF. 2022. The 5th instar Nymph of Picromerus lewisi: predation responses to different instars of Mythimna seperata. Chin Agri Sci Bull. 38(3):116–120.
- Tang YT, Guo Y, He GW, Liu CX, Chen HY, Zhang LS, Wang MQ. 2018. Functional responses of Picromerus lewisi Scott (Hemiptera: Pentatomidae) attacking Mythimna separata (Walker) (Lepidoptera: Noctuidae). Chin J Biol Control. 34(6):825–830.
- Wang Y, Wang MQ, Zhang HM, Zhao XQ, Yi YQ, Li XY, Tang YT, Chen B, Chen FS, Zhang LS. 2019. Predation potential of adult of Picromerus lewisi (Fallou) on Larvae of Spodoptera frugiperda. Chin J Biol Control. 35(5):691–697.
- Wang Y, Zhang HM, Li XY, Yi YQ, Zhao XQ, Chen FS, Zhang LS. 2020. Predation of Picromerus lewisi Nymph on Larvae of Spodoptera frugiperda. Chin J Biol Control. 36(4):520–524.
- Yang ZY, Li XW, Jia FZ, Wang MX, Yang JY, Yi ZJ, Zhang CH. 2018. Toxic effects of four commonly-used agrochemicals on Arma chinensis and Picromerus lewisi. J Agri Biotechnol. 7(5):153–155, 158.
- Zhang D, Gao F, Jakovlic I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.