Abstract
In this study, we report the female-lineage mitochondrial genome of Xenostrobus atratus for the first time. The circular mitochondrial genome is 14,806 bp in length and contains 12 protein-coding genes, 22 transfer RNA genes, and two ribosomal RNA genes. All genes are encoded on the heavy strand. The genome composition is A + T biased (66.6%), with 25.2% A, 41.4% T, 21.7% G and 11.7% C. A Bayesian inference (BI) phylogenetic tree was constructed based on the mitochondrial genomes of X. atratus and 46 other Mytilidae species. Our results demonstrate that X. atratus and Limnoperna fortunei have distinct lineages, opposing synonymizing Xenostrobus within Limnoperna. According to this study, the validity of the subfamily Limnoperninae and genus Xenostrobus is strongly supported. However, there is still an urgent need for more mitochondrial data to decide to which subfamily X. atratus belongs.
Introduction
Xenostrobus species occupys different habitats, such as tropical mangroves, rocky substrates upstream and downstream of estuaries, and rocky intertidal zone (Colgan and da Costa Citation2013). There are eight extant species of Xenostrobus (Colgan et al. Citation2020), and only one, Xenostrobus atratus, is reported for China (Bernard Citation1993; Wang Citation1997). Due to the similarity of morphological characteristics with an invasive freshwater mussel, Limnoperna fortunei (Beu Citation2006), X. atratus used to be named Limnoperna atrata. The relationship between the two genera is still in debate. For example, Beu (Citation2006) placed the species within the Limnoperna genus, however, Colgan and da Costa (Citation2013)-based on differences in BEAST-estimated age and cox1 amino acid sequences-objected to the suggestion that Xenostrobus and Limnoperna were congeneric. Therefore, convincing evidence is needed to consider the relationships between Xenostrobus and Limnoperna. In addition, the fouling property that affects the normal growth and development of mangroves (He, Citation2002) draws considerable attention to X. atratus. Since characteristics of mitochondrial genomes are efficient and powerful in phylogenetic studies in Mytilidae (Lee et al. Citation2019; Zhang et al. Citation2019), the mitochondrial genome of X. atratus was sequenced for the first time in this study. An interesting feature of Mytilidae mtDNA is Doubly Uniparental Inheritance (DUI), which has two types of mitochondrial DNA [female-lineage type (F-type) and male-lineage type (M-type)] and M-type mitogenome is only present in the gonads of male individuals. In mytlids, the F-type and M-type mitogenomes appear to exhibit an estimated 10% difference in base sequences (Fisher and Skibinski, Citation1990). DUI species were found not only in marine mussels but also in freshwater mussels (Soroka, Citation2020). There are more than 100 species of bivalve mollusks known to have DUI (Zouros, Citation2020).
Materials and methods
Mussel materials and DNA extraction
The specimen was collected from Qingdao, Shandong Province, China (119°58′12″N, 35°52′48″E) on 4 February 2022 and was stored in the Marine Biological Museum, Chinese Academy of Sciences (http://www.qdio.cas.cn/, Yongqiang Wang, [email protected], under the voucher number MBM287353). Genomic DNA was extracted from the adductor muscle by CTAB method (Doyle and Doyle Citation1987).
Genome sequencing, assembly and annotation
The X. atratus DNA library was sequenced by Personalbio Biotechnology Co., Ltd (Shanghai, China) using an Illumina NovaSeq 6000 with an average insert size of 400 bp, which was constructed using TruSeqTM Nano DNA Sample Pre Kit. Approximately 3.98 GB of raw data for X. atratus were generated with 150 bp paired-end reads. Adapters were removed by Adapter Removal v.2 (Schubert et al. Citation2016). Clean data was assembled de novo using A5-miseq v20150522 (Coil et al. Citation2015) and SPAdesv3.9.0 (Bankevich et al. Citation2012). We determined the location of genes using the MITOS web server (http://mitos2.bioinf.uni-leipzig.de/index.py). The annotated sequence has been submitted to GenBank with Accession no. OM001008 (F type).
Phylogenetic analysis
A Bayesian inference (BI) phylogenetic tree was conducted using sequence data from the concatenated sets of 12 PCGs and 2 rRNAs of the X. atratus mitogenome and 46 Mytilidae species previously published, with Crassostrea gigas (Ostreidae) and Anadara sativa (Arcidae) as outgroups. The best substitution model for nucleotide sequences selected by Modelfinder (Kalyaanamoorthy et al. Citation2017) was GTR + F + G. BI tree was established using software MrBayes v.3.2.6 (Ronquist et al. Citation2012), with 2,000,000 generations and discarding the first 25% as burnin.
Results
The mitochondrial genome of X. atratus is a closed-circular DNA molecule with a length of 14,806 bp, containing 12 protein-coding genes (PCGs), including 7 subunits of NADH dehydrogenase (nad1-6 and nad4l), 3 subunits of cytochrome c oxidase (cox1-3), one subunits of ATPase (atp6), cytochrome b (cytb), 22 transfer RNA genes (tRNAs) and 2 ribosomal RNA genes (12S rRNA and 16S rRNA). The total length of the non-coding regions is 449 bp, accounting for 3.03% of the mitochondrial genome. No atp8 gene was found by comparison with closely related species. Song et al. (Citation2009) hypothesized that the absence of gene atp8 in some species resulted from adaptation to environmental conditions, but, this coding gene has been detected in many fresh-water and marine bivalves. This might be explained by the inherent structural properties (such as short length) and extreme variability of atp8 among bivalves that hider the proper detection and annotation of this gene (Breton et al. Citation2010; Gaitán-Espitia et al. Citation2016).
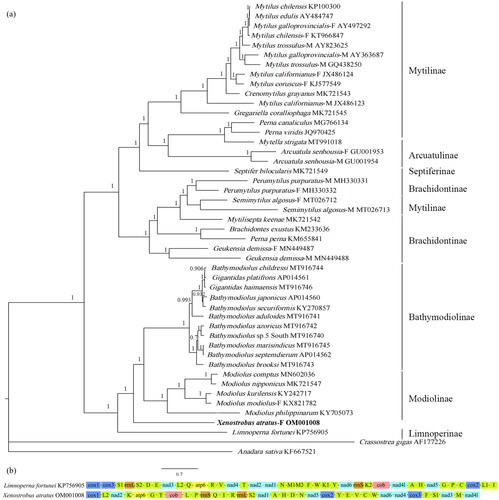
The phylogenetic tree divides the 46 Mytilidae species into 2 clades (), one of which, clusters X. atratus with Bathymodiolinae, Modiolinae, and Limnoperninae.
Figure 1. (a) The Bayesian inference phylogenetic tree for Xenostrobus atratus and other Mytilidae species, with numbers next to the node are support values; (b) Mitochondrial genome arrangement of Xenostrobus atratus and Limnoperna fortunei. The -F represents female-lineage mitochondrial genome and -M represents male-lineage mitochondrial genome.
Dicussion
The validity of the subfamily Limnoperninae has been doubted and the genus Limnoperna was sometimes placed under Arcuatulinae (Huber Citation2010). In this study, L. fortunei clusters with X. atratus, Bathymodiolinae and Modiolinae, constituting one of the two main clades in Mytilidae, and not with Arcuatulinae species. Consequently, it is more likely that Limnoperninae is valid. Lee et al. (Citation2019) proposed that Limnoperninae is sister to (Bathymodiolinae + Modiolinae) based on a mitochondrial genome phylogeny, but the newly sequenced species, X. atratus, takes its position and clusters with (Bathymodiolinae + Modiolinae) in this study, and they are then sister to Limnoperninae (posterior probability = 1). In particular, X. atratus and L. fortunei are closely related but have distinct lineages with long phylogenetic branch lengths, longer than most of the other closely related species within the Bathymodiolinae and Modiolinae, opposing the synonym of the two genera raised by Beu (Citation2006). Additionally, since gene orders among low-level taxonomic species are highly conserved and have been proven to be an effective phylogenetic tool in Mollusca (Boore and Brown, Citation1998; Ghiselli et al. Citation2021), the very large difference in the PCG order and tRNA arrangement between X. atratus and L. fortunei provides further evidence for supporting the validity of genus Xenostrobus (). Nonetheless, it’s still too early to decide which subfamily X. atratus belongs to based on current studies. Additional molecular data are needed to solve this controversy and deeper relationships among Mytilidae species.
Ethical approval
This study does not need ethical approval or permissions to collect, handling, and transport of the samples.
Author contributions
Houmei Li conducted the investigation, data curation, and drafting of the paper. Chenghua Li, Peizhen Ma, and Haiyan Wang contributed to the analysis and interpretation of data for the work. Zhen Zhang was involved in charge of the methodology and the final approval of the version to be published. All authors contributed to the article and approved the submitted version.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under accession no.OM001008. The associated BioProject, SRA and Bio-Sample numbers are PRJNA808856, SRR18136193, and SAMN26116627 respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its application to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernard FR, Cai YY, Morton B. 1993. A catalogue of the living marine Bivalve molluscs of China. Hong Kong: Hong Kong University Press.
- Beu AG. 2006. Marine Mollusca of oxygen isotope stages of the last 2 million years in New Zealand. Part 2. Biostratigraphically useful and new Pliocene to recent bivalves. J Roy Soc New Zeal. 36(4):151–338.
- Boore JL, Brown WM. 1998. Big trees from little genomes: mitochondrial gene order as a phylogenetic tool. Curr Opin Genet Dev. 8(6):668–674.
- Breton S, Stewart DT, Hoeh WR. 2010. Characterization of a mitochondrial ORF from the gender-associated mtDNAs of Mytilus spp. (Bivalvia: Mytilidae): Identification of the “missing” ATPase 8 gene. Mar Geonomics. 3(1):11–18.
- Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31(4):587–589.
- Colgan DJ, da Costa P. 2013. Invasive and non-invasive lineages in Xenostrobus (Bivalvia: Mytilidae). Molluscan Res. 33(4):272–280.
- Colgan DJ, Willan RC, Kirkendale LA. 2020. A genetic assessment of the taxonomic status of New Zealand mussels of the genus Xenostrobus Wilson, 1967. N Z J Mar Freshwater Res. 54(2):271–285.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Fisher C, Skibinski DOF. 1990. Sex-biased mitochondrial DNA heteroplasmy in the marine Mussel mytilus. Proc Biol Sci. 242(1305):149–156. http://www.jstor.org/stable/76476
- Gaitán-Espitia JD, Quintero-Galvis JF, Mesas A, D’Elía G. 2016. Mitogenomics of southern hemisphere blue mussels (Bivalvia: Pteriomorphia): Insights into the evolutionary characteristics of the Mytilus edulis complex. Sci Rep. 6(1):26853.
- Ghiselli F, Gomes-dos-Santos A, Adema CM, Lopes-Lima M, Sharbrough J, Boore JL. 2021. Molluscan mitochondrial genomes break the rules. Phil Trans R Soc B. 376(1825):20200159.
- He BY. 2002. Research on the ecology of the fouling fauna communities in mangrove. Guangxi Sci. 9(2):133–137.
- Huber M. 2010. Compendium of bivalves. A full-color guide to 3,300 of the World’s Marine Bivalves. A status on Bivalvia after 250 years of research. Hackenheim: ConchBooks.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Lee Y, Kwak H, Shin J, Kim SC, Kim T, Park JK. 2019. A mitochondrial genome phylogeny of Mytilidae (Bivalvia: Mytilida). Mol. Phylogenet. Evol. 139:106533.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Schubert M, Lindgreen S, Orlando L. 2016. AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res Notes. 9(1):88.
- Song WT, Gao XG, Li YF, Liu WD, Liu Y, He CB. 2009. Comparison of mitochondrial genomes of bivalves. Hereditas. 31(11):1127–1134.
- Soroka M. 2020. Doubly uniparental inheritance of mitochondrial DNA in freshwater mussels: history and status of the European species. J Zool Syst Evol Res. 58(2):598–614.
- Wang ZR. 1997. Fauna sinica phylum molludca order mytiloida. Beijing: Science Press; p. 206–208.
- Zhang Z, Ma PZ, Hu LS, Liu YM, Wang HY. 2019. The complete mitochondrial genome of a marine mussel, Modiolus comptus (Mollusca: Mytilidae), and its phylogenetic implication. Mitochondrial DNA Part B-Res. 4(2):4057–4058.
- Zouros E. 2020. Doubly uniparental inheritance of mitochondrial DNA: might it be simpler than we thought? J Zool Syst Evol Res. 58(2):624–631.
