Abstract
Daphne pseudomezereum A. Gray var. koreana (Nakai) Hamaya is a shrub distributed in high mountains in Japan and Korea and is used as a medicinal plant. The complete chloroplast genome of D. pseudomezereum var. koreana is 171,152 bp long with four subregions consisting of a large single-copy region (84,963 bp), a small single-copy region (41,725 bp), and a pair of inverted repeats (2739 bp). The genome includes 139 genes (93 protein-coding genes, eight rRNAs, and 38 tRNAs). Phylogenetic analyses show that D. pseudomezereum var. koreana is nested within the Daphne clade in the narrow sense and that it forms a distinct lineage.
Introduction
Daphne pseudomezereum A. Gray var. koreana (Nakai) Hamaya (Bull. Tokyo Univ. Forest. 55: 72, 1959) is a shrub distributed in high mountains in Japan and Korea (Oh and Hong Citation2015). As a member of Thymelaeaceae, which includes many ecologically important species for ornamentals, timber, papermaking, and medicine (Herber Citation2003), D. pseudomezereum can also be used as a medicinal plant for chronic skin diseases and rheumatism (Konishi et al. Citation1993). Plants of D. pseudomezereum var. koreana have rarely been collected, as they are uncommon. For this reason, little is known about the species. Detailed and accurate information on its phylogenetic position and better knowledge of the genome of the species will be useful for understanding its genetic diversity and for evaluating its phytochemical composition for medicinal purposes.
Materials and methods
The sample of D. pseudomezereum var. koreana () were collected in Sangye-ri, Okgye-myeon, Gangneung-si, Gangwon-do, Republic of Korea (37°32′48″N, 128°51'31″E). A voucher specimen was deposited in the Daejeon University Herbarium (TUT: https://www.dju.ac.kr/biosci/depart/profileView.do?mi=2253, contact person: Sang-Hun Oh, [email protected]) under the voucher number Lee 7891. Total DNA was isolated from fresh leaves of the species using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). A small portion (3 ul) of the extracted DNA was run in 1% agarose gel to examine the quality. The concentration of DNA was measured with an Invitrogen Qubit fluorometer. An amount of 1 microgram of DNA was used to reconstruct the sequencing library. We used an Illumina TruSeq Nano DNA Library Preparation Kit (Illumina, San Diego, CA) following the manufacturer’s recommendations. The sequencing library was analyzed using NovaSeq6000 at Macrogen Inc., Korea. The resulting 7.34-Gbp raw sequences were filtered using the Trimmomatic tool (v0.33) (Bolger et al. Citation2014). The chloroplast genome was de novo assembled with Velvet v1.2.10 (Zerbino and Birney Citation2008) and gaps were closed using GapCloser v1.12 (Zhao et al. Citation2011). The genome sequence was confirmed by aligning all raw reads against the assembled genome using BWA v0.7.17 and SAMtools v1.9 (Li et al. Citation2009; Li Citation2013) in the environment of Genome Information System (GeIS; https://geis.infoboss.co.kr/). Geneious Prime® v2020.2.4 (Biomatters Ltd., Auckland, New Zealand) was used for annotation based on the Daphne genkwa chloroplast (MT754180) (Yoo et al. Citation2021) and read coverage depth map (Supplementary Figure 1). A circular map of the chloroplast genome () and a schematic map of the cis- and trans-splicing genes () were generated by CPGView (Liu et al. Citation2023).
Figure 1. Photograph of Daphne pseudomezereum var. koreana. (A) Habit. (B) Flowers. Photo credit: Jae-Jin Lee.

Figure 2. Circular map of the complete chloroplast genome of Daphne pseudomezereum var. koreana. The center of the map indicates the name of the species and the length of its chloroplast genome. Going outward, the first circle shows LSC, SSC, IRa, and IRb with their length. The second circle displays the GC ratio depicted as the proportion of the shaded parts of each section. The third circle shows the gene names with the colors based on their functional categories provided in the lower left of the circular map. Genes inside the circle are transcribed in a clockwise direction, and those outside are in a counterclockwise direction.
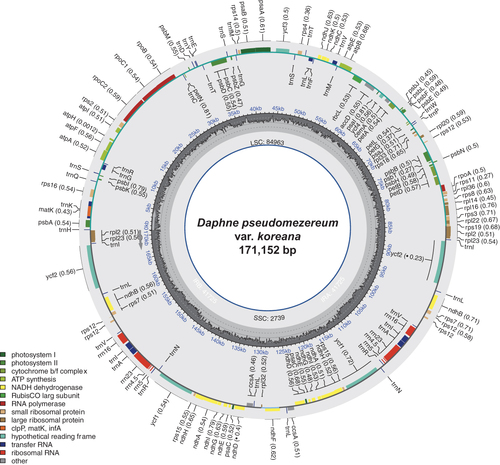
Figure 3. Phylogenetic analyses of 35 whole chloroplast genomes of Daphne and closely related species with Pimelea and Gonystylus as outgroups using the maximum likelihood (ML) and Bayesian inference (BI) methods. The following sequences are used: EU849490 (unpublished), MN201546 (Könyves et al. Citation2019), MN511715 (Qian et al. Citation2021), MN756675 (He et al. Citation2021), MT627479 (Lee et al. Citation2022), MT648376 (Lee et al. Citation2022), MT754180 (Yoo et al. Citation2021), MT790358 (Liang et al. Citation2020), MW246157 (unpublished), MW246160 (unpublished), MW393701 (unpublished), MW393702 (unpublished), NC_035896 (Cho et al. Citation2018), NC_038153 (Foster et al. Citation2018), NC_042714 (Yun et al. Citation2019), NC_042950 (Yan et al. Citation2019), NC_044085 (Yan et al. Citation2019), NC_045890 (Qian et al. Citation2020), NC_045891 (unpublished), NC_046389 (Qian and Zhang Citation2019), NC_056317 (unpublished), NC_056320 (unpublished), NC_057148 (Yan et al. Citation2021), NC_057149 (unpublished), NC_057996 (He et al. Citation2021), NC_057997 (He et al. Citation2021), NC_057998 (He et al. Citation2021), NC_057999 (He et al. Citation2021), NC_058000 (He et al. Citation2021), NC_058916 (Kim et al. Citation2021), NC_060495 (Lee et al. Citation2022), NC_060562 (Lee et al. Citation2022), NC_060829 (unpublished), NC_062858 (unpublished), ON244034 (this study). The phylogenetic tree was drawn based on the ML tree. The numbers above the branches indicate bootstrap support values of ML and the posterior probability from BI.
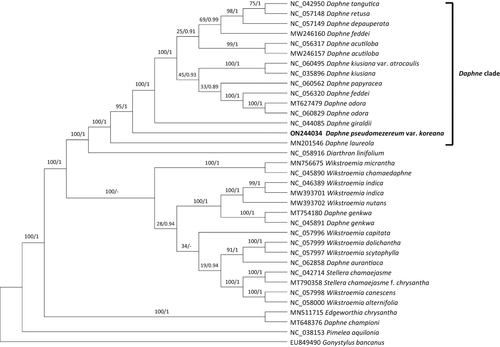
In total, 35 whole chloroplast genomes of Daphne and its closely related species (Herber Citation2003) were included in phylogenetic analyses using the maximum likelihood (ML) and Bayesian inference (BI) methods to infer the phylogenetic position of the chloroplast genome of D. pseudomezereum var. koreana. Pimelea and Gonystylus were used as outgroups. A heuristic search was conducted with nearest-neighbor interchange branch swapping, the Tamura-Nei model, and uniform rates among sites to construct the ML phylogenetic tree with default values for other options using MEGA X (Kumar et al. Citation2018). A bootstrap analysis with 1000 pseudoreplicates was also conducted with the same search options. The BI tree was constructed using MrBayes v3.2.6 (Ronquist et al. Citation2012). The GTR model with gamma rates was used as a molecular model. A Markov-chain Monte Carlo algorithm was employed for 1,000,000 generations, sampling trees every 200 generations, with four chains running simultaneously.
Results
The chloroplast genome of D. pseudomezereum var. koreana (GenBank accession: ON244034) is 171,152 bp long (). The overall GC ratio is 36.5%. The genome has four subregions consisting of 84,963 bp of a large single-copy region (LSC; 49.6%) and 2739 bp of a small single-copy (SSC; 1.6%) region separated by 41,725 bp of each of two inverted repeat regions (IRs; 48.8%). It contains 139 genes (93 protein-coding genes, eight rRNAs, and 38 tRNAs); 28 genes (16 protein-coding gene, four rRNAs, and eight tRNAs) are duplicated in IR regions ().
The phylogenetic tree showed that D. pseudomezereum var. koreana is nested within the Daphne clade, forming a distinct lineage within the clade (). It was sister to a large subclade that includes most species of Daphne, such as D. giraldii and D. tangutica ().
Discussion and conclusion
The chloroplast genome of D. pseudomezereum var. koreana exhibits long IR regions and a short SSC region compared to a typical chloroplast genome of angiosperms (Palmer et al. Citation1988; Cosner et al. Citation2004). The expansion of IR associated with the shortening of SSC is also found in other species of Thymelaeaceae, such as Daphne kiusiana (Cho et al. Citation2018) and Daphne laureola (Könyves et al. Citation2019) as well as Aquilaria sinensis (Wang et al. Citation2016), Stellera chamaejasme (Yun et al. Citation2019), and Wikstroemia chamaedaphne (Qian et al. Citation2020). It appears that the events of IR expansion and contraction should have occurred multiple times within Thymelaeaceae, and more chloroplast genomes for various taxa are needed to understand the pattern of chloroplast evolution in the family.
The phylogenetic tree indicates that the position of newly determined chloroplast genome of D. pseudomezereum var. koreana is consistent with morphology (Oh and Hong Citation2015; Lee J-J and Oh Citation2017)), placed within the core Daphne clade. The phylogenetic analysis suggests that the Daphne should be narrowly redefined to indicate the core Daphne clade (), as the traditional circumscription of the genus Daphne (Herber Citation2003; Wang et al. Citation2007; Oh and Hong Citation2015) is polyphyletic. It also suggests that Wikstroemia should expand to include D. genkwa, D. aurantiaca, and Stellera chamaejasme and that D. championi is sister to Edgeworthia chrysantha (). Our results, consistent with previous analyses (Yoo et al. Citation2021; Lee SY et al. Citation2022), indicate that further detailed systematic studies of Daphne and its closely related groups with more taxon sampling are needed. This report contributes to the understanding of the chloroplast genetic information of D. pseudomezereum var. koreana to provide additional data for future research to reconstruct the evolutionary relationships and for the establishment of a sound classification system of Daphne and its closely related groups and ultimately to develop molecular markers for the medicinal plant.
Ethical approval
The authors declare that there were no ethical or legal violations when obtaining the study materials and performing the research. The species used in this study is not listed on the IUCN Red List, and the sample was legally collected in accordance with guidelines stipulated in national and international regulations. The materials were collected in a location that was not designated as a protected area in Korea. No ethical approval or permission from the Institutional Review Board (IRB) of Daejeon University was required for this study.
Authors’ contributions
S.C. Yoo, J. Park, and S.H. Oh contributed to the data concept, design, analysis, interpretation, and drafting of this paper. S.C. Yoo contributed to the sampling, laboratory tasks, and writing of an early version of the manuscript. J. Park assembled and analyzed the genomic data and contributed to the writing of a draft of the manuscript. S.H. Oh coordinated the project, obtained financial support, generated the initial ideas, and wrote and edited a draft of the manuscript. All authors agreed on the contents of the manuscript and all are accountable for all aspects of the work.
Supplemental Material
Download MS Power Point (101.4 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The chloroplast genome sequence can be accessed via accession number ON244034 in GenBank of NCBI at https://www.ncbi.nlm.nih.gov. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA835716, SAMN28108557, and SRR19117239, respectively.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Cho W-B, Han E-K, Choi G, Lee J-H. 2018. The complete chloroplast genome of Daphne kiusiana, an evergreen broad-leaved shrub on Jeju Island. Conservation Genet Resour. 10(1):103–106.
- Cosner ME, Raubeson LA, Jansen RK. 2004. Chloroplast DNA rearrangements in Campanulaceae: phylogenetic utility of highly rearranged genomes. BMC Evol Biol. 4:27.
- Foster CSP, Henwood MJ, Ho SYW. 2018. Plastome sequences and exploration of tree-space help to resolve the phylogeny of riceflowers (Thymelaeaceae: pimelea). Mol Phylogenet Evol. 127:156–167.
- He L, Zhang Y, Lee SY. 2021. Complete plastomes of six species of Wikstroemia (Thymelaeaceae) reveal paraphyly with the monotypic genus Stellera. Sci Rep. 11(1):13608.
- Herber BE. 2003. Thymelaeaceae. In: Kubitzki K, Bayer C, editors. The families and genera of vascular plants. Vol. 5. New York: Springer-Verlag.
- Kim S-T, Oh S-H, Park J. 2021. The complete chloroplast genome of Diarthron linifolium (Thymelaeaceae), a species found on a limestone outcrop in eastern Asia. Korean J Pl Taxon. 51(4):345–352.
- Konishi T, Wada S, Kiyosawa S. 1993. Constituents of the leaves of Daphne pseudo-mezereum. Yakugaku Zasshi. 113(9):670–675.
- Könyves K, Yooprasert S, Culham A, David J. 2019. The complete plastome of Daphne laureola L. (Thymelaeaceae). Mitochondrial DNA B Resour. 4(2):3364–3365.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Lee J-J, Oh S-H. 2017. A comparative morphological study of Thymelaeacae in Korea. Korean J Pl Taxon. 47(3):207–221.
- Lee SY, Xu K-W, Huang C-Y, Lee J-H, Liao W-B, Zhang Y-H, Fan Q. 2022. Molecular phylogenetic analyses based on the complete plastid genomes and nuclear sequences reveal Daphne (Thymelaeaceae) to be non-monophyletic as current circumscription. Plant Divers. 44(3):279–289.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Liang C, Xie J, Yan J. 2020. The complete chloroplast genome sequence of Stellera chamaejasme f. chrysantha (Thymelaeaceae). Mitochondrial DNA B Resour. 5(3):3251–3252.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. (in press).
- Oh S-H, Hong H-P. 2015. Thymelaeaceae. In: Flora of Korea Editorial Committee, editor. Flora of Korea. Vol. 5b. Rosidae: Elaeagnaceae to Sapindaceae; Incheon: National Institute of Biological Resources.
- Palmer JD, Jansen RK, Michaels HJ, Chase MW, Manhart JR. 1988. Chloroplast DNA variation and plant phylogeny. Ann Missouri Botan Garden. 75(4):1180–1206.
- Qian S-J, Zhang Y-H. 2019. Characterization of the complete chloroplast genome of a medicinal plant, Wikstroemia indica (Thymelaeaceae). Mitochondrial DNA B Resour. 5(1):83–84.
- Qian S-J, Zhang Y-H, Lee SY. 2021. Comparative analysis of complete chloroplast genome sequences in Edgeworthia (Thymelaeaceae) and new insights into phylogenetic relationships. Front Genet. 12:643552.
- Qian S-J, Zhang Y-H, Li G-D. 2020. The complete chloroplast genome of a medicinal plant, Wikstroemia chamaedaphne (Thymelaeaceae). Mitochondrial DNA B Resour. 5(1):648–649.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Wang Y, Gibert M, Mathew B, Brickell C, Nevling L. 2007. Thymelaeaceae. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 13. (Clusiaceae through Araliaceae). Beijing and Missouri Botanical Garden, St. Louis: Science Press.
- Wang Y, Zhan D-F, Jia X, Mei W-L, Dai H-F, Chen X-T, Peng S-Q. 2016. Complete chloroplast genome sequence of Aquilaria sinensis (Lour.) Gilg and evolution analysis within the Malvales order. Front Plant Sci. 7:280.
- Yan F, Tao X, Wang Q-L, Juan ZY, Zhang C-M, Yu HL. 2019. The complete chloroplast genome sequence of the medicinal shrub Daphne giraldii Nitsche. (Thymelaeaceae). Mitochondrial DNA B Resour. 4(2):2685–2686.
- Yan F, Wang Q-L, Zhang Y-J, Zhang C-M, Chen Y. 2019. The complete chloroplast genome sequence of medicinal plant, Daphne tangutica Maxim. (Thymelaeaceae). Mitochondrial DNA B. 4(1):1776–1777.
- Yan F, Zhang C-Y, Wang Q-L, Wang J-D, Wang H-P, Xu T, Wang E-J, Hou L-Y. 2021. Characterization of the complete chloroplast genome sequence of Daphne retusa Hemsl. (Thymelaeaceae), a rare alpine plant species in northwestern China. Mitochondrial DNA B Resour. 6(8):2139–2141.
- Yoo S-C, Oh S-H, Park J. 2021. Phylogenetic position of Daphne genkwa (Thymelaeaceae) inferred from complete chloroplast data. Korean J Pl Taxon. 51(2):171–175.
- Yun N, Park J, Oh S-H. 2019. The complete chloroplast genome of the traditional medicinal plant Stellera chamaejasme L. (Thymelaeaceae). Mitochondrial DNA B. 4(1):1796–1797.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12(S14):S2.
