Abstract
The lawn cutworm, Spodoptera depravata, is one of the most important pests that causes economic damage to grass crops. This study reports the complete mitochondrial genome of an S. depravata sample collected in China. The genome is a circular molecule 15,460 bp in length with an overall A + T content of 81.6%. It contains 13 protein-coding genes, 22 transfer RNA genes, and two ribosomal RNA genes. The gene content and organization of the mitogenome of S. depravata are identical to those of other Spodoptera species. Maximum-likelihood phylogenetic analysis based on mitogenomes showed a close evolutionary relationship between S. depravata and S. exempta. This study provides new molecular data for the identification and further phylogenetic analyses of Spodoptera species.
Introduction
The lawn cutworm, Spodoptera depravata (Butler, 1879), is a major lepidopteran pest exclusively distributed in northeastern Asian countries, such as China, Japan, and the Republic of Korea (Oku et al. Citation1978; Kang et al. Citation2004). The larvae of S. depravata attack the linear leaves of Gramineae crops, resulting in yellowish grasses and brown patches on the ground (Kang et al. Citation2004). Previous studies on this pest have focused on ecological research, including their flight activity, reproductive capacity, life cycle, and control methods (Saito Citation2000; Qian et al. Citation2003; Huang et al. Citation2004; Huang and Xu Citation2004; Zhou et al. Citation2008). To date, few molecular biological studies of this pest have been conducted. As a member of the Spodoptera genus of moths, which includes pest insects comprising approximately 30 species, S. depravata causes severe damage to multiple agricultural crop species globally (Kergoat et al. Citation2021). The comprehensive identification and phylogenetic analysis of this genus will help clarify the relationships between different species and help prevent potential crop damage. However, mitochondrial genome data have been published for only five species of Spodoptera, and the genetic relationship between species requires more in-depth investigation. In this study, the complete mitochondrial genome of S. depravata () was sequenced.
Figure 1. Species reference image of Spodoptera depravata. The image is referenced from the Bold Systems (Ratnasingham and Hebert Citation2007). Sample ID is SDNU-INS-04111. This figure is attribution noncommercial sharing. http://v3.boldsystems.org/index.php/Taxbrowser_Taxonpage?taxon=Spodoptera+depravata&searchTax=.

Materials
The S. depravata sample used in this study was a moth collected from Changchun (43°48′38″N, 125°24′14″E), Jilin Province, China in 2020. The specimen was initially identified as belonging to the genus Spodoptera, based on its morphological appearance (Chen Citation1999), and was further identified as S. depravata based on the mitochondrial cytochrome oxidase subunit 1 (COI) sequence (Folmer et al. Citation1994). A nucleotide BLAST search in GenBank showed that the COI sequence of the specimen had 99.51–100.00% sequence similarity with the COI sequence of S. depravata. The phylogenetic tree constructed using the neighbor-joining method also showed that the specimen was grouped with S. depravata samples, including three previously published samples (Watabiki et al. Citation2013; Kergoat et al. Citation2021). The voucher specimen and DNA were deposited at the Agricultural Genomics Institute in Shenzhen, China (https://www.agis.org.cn/, contact Lei Zhang, [email protected]) under voucher number AGIS-SD-202011.
Methods
Genomic DNA was extracted using the DNeasy Blood and Tissue Kit (Cat. no. 69506; Qiagen, Hilden, Germany). DNA quality and concentration were determined using a NanoDrop One UV-Vis spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). A total of 0.5 μg of genomic DNA was used to construct a library with a 350 bp insert size. The library was sequenced on the NovaSeq 6000 platform (Illumina, San Diego, CA, USA) with 150 bp paired-end reads, resulting in a total of 144,834,262 reads (the percentage of reads with ≥ Q30 score was 95.88%). Moreover, we further evaluated the sequencing depth of mitochondrial genome using mosdepth software (Pedersen and Quinlan Citation2018).
The complete mitochondrial genome sequence was assembled using the software NOVOPlasty v2.5.6 (Dierckxsens et al. Citation2017), and the mitogenome was annotated using MITOS2 (http://mitos2.bioinf.uni-leipzig.de/index.py) (Donath et al. Citation2019). A mitochondrial genome map of S. depravata was generated using OGDRAW v1.3.1 software (Greiner et al. Citation2019). Multiple mitochondrial genome sequence alignments were conducted using MAFFT v7.455 (Katoh and Standley Citation2013). A phylogenetic tree was constructed based on the maximum-likelihood method using the best-fit model (GTR + F+I + G4) estimated using IQ-TREE v1.6.10 (Nguyen et al. Citation2015). One thousand bootstrap replicates were used, and the tree was visualized using iTol v6.5.2 (https://itol.embl.de/).
Results
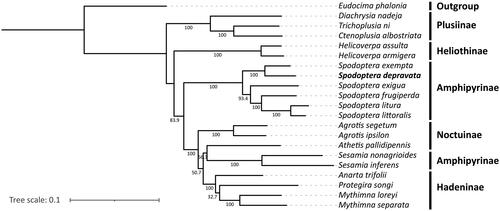
The mitogenome of S. depravata (GenBank Accession Number OK053108) is a circular molecule 15,460 bp in length, with an A + T content of 81.6%. The mitogenome consensus sequence average coverage was 5866× (Figure S1). However, the differences in length between species are primarily in the non-coding regions. The mitogenome encodes a standard set of 37 genes, including 13 protein-coding genes (PCGs), two ribosomal RNAs, and 22 transfer RNAs, and a control region (). All 13 PCGs begin with ATN codons, except for cox1, which begins with CGA. Most PCGs are terminated with TAA or TAG stop codons, except for three genes (cox1, cox2, and nad4), which have an incomplete stop codon (T), as seen in S. exempta and S. littoralis. In contrast, five genes (nad2, cox1, cox2, nad3, and nad4) have incomplete stop codons (T) in S. exigua, S. frugiperda, and S. litura. Twenty-two complete mitogenomes (21 lepidopteran species and one species from the family Erebidae) were used for phylogenetic analysis (). The phylogenetic tree showed that all six Spodoptera species formed a clade, in which four species were clustered into one branch ([{S. littoralis + S. litura} + S. frugiperda] + S. exigua), which is consistent with the findings of previous studies (Li, Wang, Xiao, et al. Citation2021). The other branch consisted of S. depravata and S. exempta, indicating the close evolutionary relationship between S. depravata and S. exempta.
Figure 2. Gene map of S. depravata mitochondrial genome. Genes belonging to different functional groups are color-coded. Genes are shown outside and inside the outer circle are transcribed counterclockwise and clockwise, respectively. The grey in the inner circle corresponds to the GC% along the mitochondrion.
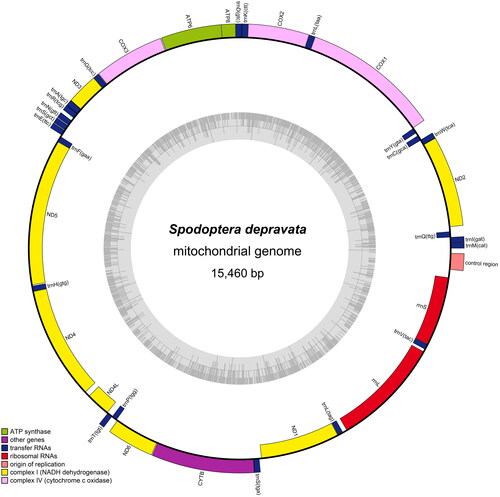
Figure 3. Phylogenetic tree of 21 lepidopteran species based on their mitochondrial genomes. Eudocima phalonia from the family Erebidae was used as the outgroup. The following sequences were used: Eudocima phalonia NC_032382.1 (Seo et al. Citation2019), Diachrysia nadeja MT916722.1 (Gao et al. Citation2021), Trichoplusia ni MK358044.1 (Zhao et al. Citation2022), Ctenoplusia albostriata NC_053742.1 (Xue et al. Citation2019), Helicoverpa assulta NC_014668.1 (Seo et al. Citation2019), Helicoverpa armigera NC_014668.1 (Yin et al. Citation2010), Spodoptera exempta NC_054179.1 (Li, Wang, Xiao, et al. Citation2021), Spodoptera exigua JX316220.1 (Wu et al. Citation2013), Spodoptera frugiperda MN385596.1 (Li, Wang, Xiao, et al. Citation2021), Spodoptera litura NC_022676.1 (Wan et al. Citation2013), Spodoptera littoralis MT816470.1 (Li, Wang, and Zhang Citation2021), Agrotis segetum KC894725.1 (Wu et al. Citation2014), Agrotis ipsilon KF163965 (Zhao et al. Citation2022), Athetis pallidipennis NC_046525.1 (Li, Wang, and Zhang Citation2021), Sesamia nonagrioides CM030612.1 (Muller et al. Citation2021), Sesamia inferens JN039362.1 (Wu et al. Citation2019), Anarta trifolii NC_046049.1 (Li, Wang, and Zhang Citation2021), Protegira songi NC_034938.1 (Guo et al. Citation2017), Mythimna loreyi NC_057500.1 (Nam et al. Citation2020), Mythimna separata KM099034.1 (Li, Wang, and Zhang Citation2021).
Discussion and conclusion
We assembled and annotated the first complete mitochondrial genome sequence of S. depravata, which is 15,460 bp in length. It is longer than the mitogenomes of the other five published Spodoptera species, which range from 15,365 to 15,457 bp (Wan et al. Citation2013; Wu et al. Citation2013; Seo et al. Citation2019; Li, Wang, and Zhang Citation2021; Li et al. Citation2021). The gene composition and gene order are the same for all six Spodoptera mitogenomes and are identical to those of the majority of lepidopteran species (Wan et al. Citation2013). We also conducted a phylogenetic analysis of the Spodoptera species. It provides useful information and lays a foundation for the subsequent genetic evolution and phylogenetic analyses of Spodoptera species.
Ethical approval
Spodoptera depravata is a common pest on grass crops in China. Therefore, no ethical approval or other relevant permission can be provided for the study.
Authors’ contributions
Xinyue Liang and Lei Zhang collected and identified the specimens; Xinyue Liang and Zaiyuan Li analyzed the data; Xinyue Liang wrote the manuscript; Lei Zhang and Zaiyuan Li revised the manuscript. Yutao Xiao conceived and designed the project, and finally approved the version to be published. All authors agree to be accountable for all aspects of the work.
Supplemental Material
Download PDF (302.4 KB)Supplemental Material
Download MS Word (673.4 KB)Disclosure statement
No potential conflict of interest is reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under accession no. OK053108. The associated BioProject, BioSample, and SRA numbers are PRJNA809185, SAMN26149759, and SRS12076762 respectively.
Additional information
Funding
References
- Chen YX. 1999. Lepidoptera: Noctuidae. [Fauna Sinica] Insect. Vol. 16. Beijing: Science Press; p. 1596
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Donath A, Jühling F, Al-Arab M, Bernhart SH, Reinhardt F, Stadler PF, Middendorf M, Bernt M. 2019. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 47(20):10543–10552.
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 3(5):294–299.
- Gao SS, Xue S, Zhang YC, Wang JS, Zhang KP. 2021. Mitochondrial genome of Diachrysia nadeja (Lepidoptera: Noctuoidea: Noctuidae) and phylogenetic analysis. Mitochondrial DNA B Resour. 6(2):406–407.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3. 1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Guo XH, Cheng MY, Hou YF, Deng TT, Zhao S, Yu HL, Xing LX. 2017. The complete mitochondrial genome of the monophagous moth Protegira songi (Lepidoptera: Noctuidae). Conservation Genet Resour. 9:277–279.
- Huang D, Xu H. 2004. Experimental population life tables of Spodoptera depravata. Jiangsu J Agri Sci. 20(3):164–168.
- Huang D, Xu Z, Li J, Dai Y. 2004. Preliminary studies on the survival and reproductive capacity of Spodoptera depravata. J Yangzhou Univ. 25(1):73–75.
- Kang YJ, Lee DW, Choo HY, Lee SM, Kweon TW, Shin HK. 2004. Biological control of Spodoptera depravata (Butler) (Lepidoptera: noctuidae) using entomopathogenic nematodes. Korean J Appl Entomol. 43:61–70.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kergoat GJ, Goldstein PZ, Le Ru B, Meagher RL, Zilli A, Mitchell A, Clamens AL, Gimenez S, Barbut J, Nègre N, et al. 2021. A novel reference dated phylogeny for the genus Spodoptera Guenée (Lepidoptera: Noctuidae: Noctuinae): new insights into the evolution of a pest-rich genus. Mol Phylogenet Evol. 161:107161.
- Li ZY, Wang WK, Xiao YT, Zhang L. 2021. Characterization of the mitochondrial genome of Spodoptera exempta (Lepidoptera: Noctuidae) from South Africa. Mitochondrial DNA B Resour. 6(2):370–372.
- Li ZY, Wang WK, Zhang L. 2021. Complete mitochondrial genome of Spodoptera littoralis (Lepidoptera: Noctuidae) from Egypt. Mitochondrial DNA B Resour. 6(2):432–434.
- Muller H, Ogereau D, Da Lage JL, Capdevielle C, Pollet N, Fortuna T, Jeannette R, Kaiser L, Gilbert C. 2021. Draft nuclear genome and complete mitogenome of the Mediterranean corn borer, Sesamia nonagrioides, a major pest of maize. G3-Genes Genomes Genet. 11(7):jkab155.
- Nam HY, Kwon M, Kim HJ, Kim J. 2020. Development of a species diagnostic molecular tool for an invasive pest, Mythimna loreyi, using LAMP. Insects. 11(11):817.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Oku T, Kobayashi T, Saito O. 1978. Studies on the ecology and control of grassland insects, VIII: notes on the phenology and some behaviors of a grass-worm, Spodoptera depravata Butler, in Morioka [Japan. ]. Bull Tohoku Natl Agric Exp Stn. 58:81–96.
- Pedersen BS, Quinlan AR. 2018. Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics. 34(5):867–868.
- Qian ZG, Shen GH, Xia XL, Luo QH, Yang L, Yuan YD. 2003. Studies on the biological character and control of Sidemia depravata (Butler). Acta Agric Shanghai. 19(03):93–96.
- Ratnasingham S, Hebert PD. 2007. bold: the Barcode of Life Data System (http://www.barcodinglife.org). Mol Ecol Notes. 7(3):355–364.
- Saito O. 2000. Flight activity of three Spodoptera spp., Spodoptera litura, S. exigua and S. depravata, measured by flight actograph. Physiol Entomol. 25(2):112–119.
- Seo BY, Lee GS, Park J, Xi H, Lee H, Lee J, Park J, Lee W. 2019. The complete mitochondrial genome of the fall armyworm, Spodoptera frugiperda Smith, 1797 (Lepidoptera; Noctuidae), firstly collected in Korea. Mitochondrial DNA B Resour. 4(2):3918–3920.
- Wan X, Kim MJ, Kim I. 2013. Description of new mitochondrial genomes (Spodoptera litura, Noctuoidea and Cnaphalocrocis medinalis, Pyraloidea) and phylogenetic reconstruction of Lepidoptera with the comment on optimization schemes. Mol Biol Rep. 40(11):6333–6349.
- Watabiki D, Yoshimatsu S, Yoshitake H, Baba Y, Uesato T, Shimatani M, Ibusuki H, Yuda T. 2013. Discrimination methods for Japanese pest species of Spodoptera Guenee (Lepidoptera: Noctuidae) lured by traps using a synthetic sex pheromone for Spodoptera exempta. JPN J Appl Entomol Zool. 57(1):19–26.
- Wu K, Yang JG, Ni YY, Liu QN. 2019. Identification and analysis of the complete mitochondrial genome of Thaumetopoea pityocampa (Lepidoptera: Notodontidae). Mitochondrial DNA B Resour. 4(2):3654–3656.
- Wu QL, Cui WX, Du BZ, Gu Y, Wei SJ. 2014. The complete mitogenome of the turnip moth Agrotis segetum (Lepidoptera: Noctuidae). Mitochondrial DNA. 25(5):345–347.
- Wu QL, Gong YJ, Gu Y, Wei SJ. 2013. The complete mitochondrial genome of the beet armyworm Spodoptera exigua (Hübner) (Lepodiptera: Noctuidae). Mitochondrial DNA. 24(1):31–33.
- Xue S, Zhang YC, Gao SS, Zhang ML. 2019. Characterisation and phylogenetic analysis of the complete mitochondrial genome of Ctenoplusia albostriata (Lepidoptera: Noctuidae: Plusiinae). Mitochondrial DNA B Resour. 4(2):3509–3510.
- Yin J, Hong GY, Wang AM, Cao YZ, Wei ZJ. 2010. Mitochondrial genome of the cotton bollworm Helicoverpa armigera (Lepidoptera: Noctuidae) and comparison with other Lepidopterans. Mitochondrial DNA. 21(5):160–169.
- Zhao JR, Zhang SP, Tang YY, Wang WZ, Tang BP, Liu QN, Yang RP. 2022. Comparative mitochondrial genome analysis of Mamestra configurata (Lepidoptera: Noctuoidea: Noctuidae) and other noctuid insects reveals conserved genome organization and phylogeny. Ann Entomol Soc Am. 115(3):304–313.
- Zhou XW, Deng JH, Gu JR, Zhang Q, Yang DF. 2008. Studies on biological characteristics and control of Sidemia depravata Bulter. J Jinling Institute Tech. 24(2):81–84.
