Abstract
Ischnura asiatica (Brauer, 1865) is a freshwater damselfly belonging to the family Coenagrionidae that is distributed across most of Korea, primarily in areas with low water flow, such as ponds and wetlands. The complete mitochondrial genome of I. asiatica was sequenced by next-generation sequencing. The circular mitochondrial genome was found to be 15,769 bp long, with of 13 protein-coding, two ribosomal RNA, and 22 transfer RNA genes (GenBank accession no. OM310774). Maximum likelihood, phylogenetic analysis showed that this species clustered with other species belonging to the family Coenagrionidae. This study contributes to the phylogeny of damselflies and other members of the family Coenagrionidae.
Ischnura asiatica (Brauer, 1865) is a freshwater invertebrate belonging to the family Coenagrionidae. It is distributed across most of Korea, primarily in areas with low water flow, such as ponds and wetlands (Lee et al. Citation2018), and is an important primary consumer in shallow lake ecosystems (Van de Meutter et al. Citation2005; Culler et al. Citation2014). This invertebrate has a relatively small body size and is prey for fishes and other invertebrates as larvae, while for birds and spiders as adults (Outomuro and Johansson Citation2015; Srivastava et al. Citation2020). This species has several morphological adaptations to local environmental characteristics and as a part of its defenses against predators. Molecular sequencing of the mitochondrial genome would contribute significantly to understanding genetic variations in this species. The phylogeny of Coenagrionidae is not well known, except for the cytochrome c oxidase subunit I gene. The complete mitochondrial genome sequence of I. asiatica has not yet been elucidated. In this study, we analyzed the mitochondrial genome of I. asiatica using next-generation sequencing. These results will be useful in further phylogenetic analyses of the family Coenagrionidae.
Ischnura asiatica specimens were collected from a tributary (Bokhacheon) of the Han River, South Korea (37°17′41.80″ N, 127°29′23.20″ E). Ischnura asiatica larvae were collected during a 30–40 min period by sweeping over the sediment surface and leaves and stems of aquatic macrophytes using a stainless-steel sampler (30 cm width, 600-µm mesh). The samples were sorted to remove plant leaves, stems, and other debris. Collected specimens were identified to species level according to Yoon (Citation1995) and Kawai and Tanida (Citation2005) and stored in the specimen museum of Dongju College (accession number: DCN-0109002, Kwang-Seuk Jeong, [email protected]).
Ischnura asiatica specimens were homogenized (using a Precellys® 24D homogenizer; Bertin, Montigny-le-Bretonneux, France), and DNA samples were prepared using a DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). Library preparation (Illumina TruSeq DNA PCR-free; Cat. No. 20015963) and assembly (SPAdes, V 3.12), and data preparation for DNA sequencing (101 bp from paired-ends on an Illumina NovaSeq 6000) was performed by Macrogen Inc. (Seoul, Korea). The genome was assembled using MEGA-X software (Kumar et al. Citation2018). We used maximum likelihood implemented in IQ-TREE software (ver. 1.6.12; Nguyen et al. Citation2015) to construct the phylogenetic tree using the protein-coding gene sequences. The annotated mitochondrial genome sequence of I. asiatica is available in the National Center for Biotechnology Information database (Data Availability Statement).
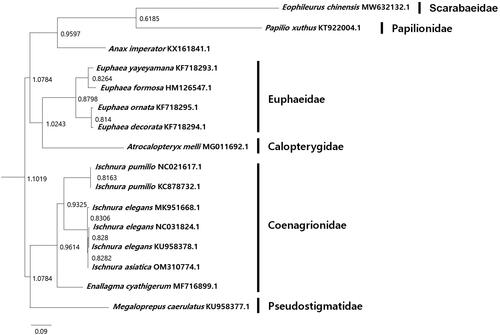
The circular mitochondrial DNA (mtDNA) of I. asiatica is 15,769 bp and consists of 13 protein-coding genes (atp6, atp8, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, cox1, cox2, cox3, and cob), two ribosomal RNA genes (rrnL, rrnS) and 22 transfer RNA genes (trnA, trnC, trnD, trnE, trnF, trnG, trnH, trnI, trnK, trnL1, trnL2, trnM, trnN, trnP, trnQ, trnR, trnS1, trnS2, trnT, trnV, trnW, and trnY), and the control region (between bps 14,789–15,769). The A + T base composition of the genome is 72.4%, and the A + T content of the genes ranges from 59.1% to 80.6%. The G + T base composition of the genome is 27.6%, and the G + C content of the genes ranges from 19.4% to 40.9%. The most common start codon was ATG, followed by ATT. The most common stop codon was TAA. Using IQ-TREE software, a phylogenetic analysis based on the mitochondrial genome of I. asiatica and similar mtDNA sequences from the order Odonata (downloaded from GenBank following a BLASTN search) showed that I. asiatica clustered with the clade of other species in the Ischnura genus with a bootstrap support value of 94%. It also revealed that the family Coenagrionidae was not monophyletic (). Considering that the nuclear genomes in the family Coenagrionidae, to which I. asiatica belongs have not been sequenced, the availability of a complete mitochondrial genome sequence for I. asiatica will be valuable for future phylogeographical studies of the family Coenagrionidae.
Recently, climate change is a major driving force in determining species distribution, and Zygoptera (damselflies) are also known to be affected (Ott Citation2010; Jung et al. Citation2016). Understanding mitochondrial genome evolution may help researchers prepare strategies for preserving or managing distribution of I. asiatica against climate change, including through accurate species identification through molecular barcoding (Rubinoff Citation2006), or investigation of their energy metabolism (Bratic and Trifunovic Citation2010).
Ethical approval
This study did not involve humans or animals. This study did not require ethical approval or permission to collect samples.
Author contributions
Jeong and Choi developed the concept and research design in the first stage of this study and undertook sample collection and preparation. Hyunbin Jo and Kwang-Seuk Jeong performed the next generation sequencing data analysis and interpretation. Based on the results, Keon-Young Jeong and Jong-Yun Choi drafted the manuscript, and Hyunbin Jo critically revised the manuscript for intellectual content. Keon-Young Jeong and Kwang-Seuk Jeong approved the final draft of the manuscript. All authors agree to be accountable for all aspects of this work.
Acknowledgements
We were indebted to Mr. Seong-Ki Kim’s effort in critical manuscript revision.
Disclosure statement
The authors declare that there is no conflict of interest regarding this study.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under accession no. OM310774. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA778563, SRR19369490, and SAMN22964840, respectively.
Additional information
Funding
References
- Bratic I, Trifunovic A. 2010. Mitochondrial energy metabolism and ageing. Biochim Biophys Acta Bioenerg. 1797(6–7):577–1342.
- Culler LE, McPeek MA, Ayres MP. 2014. Predation risk shapes thermal physiology of a predaceous damselfly. Oecologia. 176(3):653–660.
- Jung SW, Min HK, Hwang HS, Seo YJ, Bae YJ, Paek WK. 2016. Diversity of aquatic insects of taean area in South Korea, with notes on species-specific distribution. Kor J Environ Ecol. 30(1):58–70.
- Kawai T, Tanida K. 2005. Aquatic insects of Japan: manual with keys and illustrations; Tokai University Press: Kanagawa, Japan.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Lee DY, Lee DS, Bae MJ, Hwang SJ, Noh SY, Moon JS, Park YS. 2018. Distribution patterns of odonate assemblages in relation to environmental variables in streams of South Korea. Insects. 9(4):152.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Ott J. 2010. Dragonflies and climatic change – recent trends in Germany and Europe. BioRisk. 5:253–286.
- Outomuro D, Johansson F. 2015. Bird predation selects for wing shape and coloration in a damselfly. J Evol Biol. 28(4):791–799.
- Rubinoff D. 2006. Utility of mitochondrial DNA barcodes in species conservation. Conserv Biol. 20(4):1026–1033.
- Srivastava DS, Ware JL, Ngai JT, Starzomski BM, Amundrud SL. 2020. Habitat size thresholds for predators: why damselflies only occur in large bromeliads. Biotropica. 52(6):1030–1040.
- Yoon IB. 1995. Aquatic insects of Korea; Jeonghaengsa: Seoul, Korea.
- Van de Meutter F, De Meester L, Stoks R. 2005. Water turbidity affects predator–prey interactions in a fish–damselfly system. Oecologia. 144(2):327–336.