Abstract
In the present study, the complete mitochondrial genome of Lepidozona coreanica was sequenced and described. The complete mitogenome sequence of L. coreanica is 16,572 bp long and contains 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, and two ribosomal RNA (rRNA) genes. The base composition was AT biased (70.1%). The 13 PCGs of L. coreanica and the other 15 species of Polyplacophora were used for phylogenetic analysis using maximum-likelihood methods. The results showed that L. coreanica, Ischnochiton hakodadensis, and Chaetopleura apiculata are sister groups of the three lineages.
Introduction
Lepidozona coreanica (Reeve, 1847) (Polyplacophora, Chitonida, Ischnochitonidae) () is an oblong and oval-shaped marine mollusk averaging 22 mm in length and 15 mm in width, which is morphologically composed of a large muscular foot that allows it to adapt conform to irregular surfaces (Garcia-Ibanez et al. Citation2013; Owada Citation2018). In China, it is commonly found in the intertidal zone along the coast, particularly on the north coast; however, it is uncommon on the south coast. It has also been spotted in the Korean Peninsula and Japan (from Hokkaido to south of Kyushu) and plays an important role in maintaining community structure and dynamics on rocky shores (Xu et al. Citation2020). Most chitons are edible and have medicinal properties. In particular, L. coreanica, has been used as a research material for biomineralization and natural nanomaterials containing large amounts of magnetite in the form of nanoparticles (Kirschvink and Lowenstam Citation1979). The taxonomy of L. coreanica has not yet been clarified at the molecular biological level; thus, it has only provisionally been assigned to the Ischnochitonidae family (Higo Citation1999). Therefore, it is important to elucidate the taxonomy of L. coreanica based on the complete mitochondrial genome while considering the threat to population genetic diversity from coastal land reclamation and commercial exploitation.
Materials and methods
Samples used for sequencing were collected from Jingouzhai, Yantai, Shandong Province, China (37°31′52″N, 121°26′16″E). The samples were stored in 70% ethanol immediately after collection (Pu et al. Citation2017). A specimen has been deposited in the marine specimen room of Yantai University (https://lsc.ytu.edu.cn/index.htm, Jiangyong Qu, [email protected]) under the specimen number YTU-LSC-201900370015. The mitochondrial genome sequence of L. coreanica has been stored in GenBank under accession number NC_046935. Genomic DNA was extracted from animals using a mitochondrial DNA column extraction kit (Sequencing Grade, BioRab, Beijing, China), and quality control was performed on the purified DNA samples. The mitochondrial genome was sequenced using the Illumina NovaSeq 6000 sequencing platform (Shanghai BIOZERON Co., Ltd., Shanghai, China). Readings were filtered using Trimmomatic 0.39 (http://www.usadellab.org). The Q30 of the clean data was 94.3%, which could be used for subsequent compilation and analysis. The clean data were compiled using SPAdes v3.10.1 (http://bioinf.spbau.ru/spades). The MITOS software (http://mitos.bioinf.uni-leipzig.de/index.py) was used to identify protein-coding genes (PCGs), transfer RNA (tRNA), and ribosomal RNA (rRNA) genes in the mitochondrial genome. The mitochondrial genome was obtained using OrganellarGenomeDRAW (Greiner et al. Citation2019). The phylogenetic tree was constructed by maximum-likelihood (ML) in MEGA v11 (Tamura et al. Citation2021), using 13 PCGs from 17 mitogenomes (L. coreanica, other 15 species from Polyplacophora and one outgroup Haliotis discus hannai). Bootstrap values are evaluated using the bootstrap method with 1000 repetitions.
Results
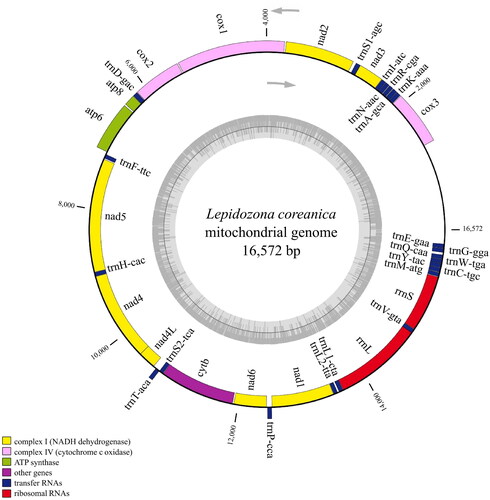
The complete mitochondrial genome of L. coreanica was 16,572 bp in length (GenBank accession number: NC_046935) and comprised 13 PCGs, 22 tRNA genes, and two rRNA genes (). The overall base composition was A 36.1%, T 34.1%, G 14.5%, and C 15.4%, with a GC content of 29.9%. Similar to the mitogenomes of other Polyplacophora species, the GC content was almost 30% (Cui et al. Citation2019; Guo et al. Citation2019). Eight of the 13 PCGs started with ATG (cox1, cox2, cox3, atp6, atp8, nad1, nad3, and nad5), two genes started with ATT (nad2 and cytb), two genes started with ATA (nad4L, and nad6), and one gene started with GTG (nad4). Seven of the 13 PCGs had stop codons TAA (nad1, nad2, nad3, atp6, cox1, cox3, and cytb), five had TAG (cox2, nad4L, nad5, nad6, and atp8), and nad4 ended with an incomplete stop codon T. Two rRNA genes, 12SrRNA and 16SrRNA, were isolated by tRNA-Val (GTA). The 22 tRNA genes were between 64 bp and 71 bp in length.
Discussion and conclusions
These mitochondrial genomes share the same gene order in terms of the relative position of PCGs; however, some tRNAs are rearranged (). The main strand encoded seven PCGs (cox1, cox2, atp6, atp8, cox3, nad3, and nad2) and 10 tRNAs (trnD, trnK, trnT, trnP, trnK, trnA, trnR, trnN, trnI, and trnS1). The remaining PCGs (nad5, nad4, nad4L, cytb, and nad1) and 12 tRNAs (trnF, trnH, trnS1, trnP, trnL1, trnL2, trnV, trnM, trnY, trnC, trnW, and trnQ) were located in the minus strand. The trnF, trnS2, trnV, trnW, and trnT tRNAs are arranged in either the minus strand or main strand. Both rRNA genes are transcribed from the minus strand, with the 16S rRNA being flanked by trnL and trnV; the 12S rRNA is located between trnV and trnM. The genome structure and gene composition were comparable to those of species of the Polyplacophora class, with the exception of , indicating that the control region was relatively stable. We interrupted the non-coding control region between trnE and cox3 in the circle mitochondrial genome and placed it in the middle of a determined linear sequence for BWA alignment, and calculated the coverage depth at each position (Additional ).
Figure 3. Mitochondrial gene orders of Polyplacophora main lineages: genes encoded in the major and minor strands are shown in the top and bottom lines, respectively. The rearrangement of genes is colored.

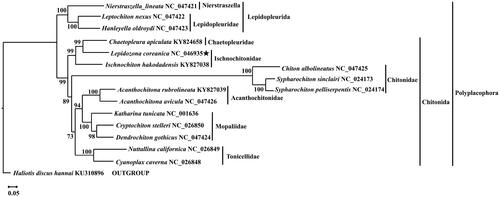
The results of the constructed phylogenetic tree showed with strong support that L. coreanica and Chaetopleura apiculata are sister groups (SH-aLRT support = 99%, ). It also showed with strong support that I. hakodadensis (Ischnochitonidae) was sister to L. coreanica (Ischnochitonidae) and Chaetopleura apiculata (Chaetopleuridae) (SH-aLRT support = 99%, ). Furthermore, the phylogenetic tree showed that Ischnochitonidae (Chitonida) was relatively distant from Nierstraszellidae (Lepidopleurida) and that the Ischnochitonidae (Chitonida), Chitonidae (Chitonida), and Chaetopleuridae (Chitonida) families were closer, which is consistent with other studies (Irisarri et al. Citation2020). These results provide new insights into the diversity of the Polyplacophora mitogenome and the evolution of chitons.
Figure 4. Phylogenetic position of L. coreanica based on 13 mitochondrial protein-coding genes. Sixteen ingroups (Polyplacophora) and one outgroup. Numbers next to each node indicate bootstrap support values. The content in parenthesis after the species name is the GenBank accession number. The following sequences were used: NC_047421 (Irisarri et al. Citation2020), NC_047422 (Irisarri et al. Citation2020), NC_047423 (Irisarri et al. Citation2020), KY824658 (Guerra et al. Citation2018), KY827038 (Cui et al. Citation2019), NC_047425 (Irisarri et al. Citation2020), NC_024173 (Veale et al. Citation2016), NC_024174 (Veale et al. Citation2016), KY827039 (Guo et al. Citation2019), NC_047426 (Irisarri et al. Citation2020), NC_001636 (Boore and Brown Citation1994), NC_026850 (Irisarri et al. Citation2020), NC_047424 (Irisarri et al. Citation2020), NC_026849 (Irisarri et al. Citation2020), NC_026848 (Irisarri et al. Citation2020), and KU310896 (Pu et al. Citation2020).
Author contributions
Conceived and designed the experiments: Lijun Wang, Jiangyong Qu, Xumin Wang, and Zhikai Xing. Performed the experiments: Donghui Sun and Zhongyu Lin. Analyzed the data: Donghui Sun, Xindong Teng, Li Xu, Lijia Qian, Xinyue Yu, Huafang Wu, Ziyi Wang, and Liming Jin. Contributed reagents/materials/analysis tools: Donghui Sun, Zhongyu Lin, and Xiumei Liu. Wrote the paper: Donghui Sun and Zhongyu Lin. Final approval of the version to be published: Lijun Wang, Jiangyong Qu, Xumin Wang, and Zhikai Xing. All authors agree to be accountable for all aspects of the work.
Ethical approval
All animal protocols have been reviewed and approved by the Experimental Animal Welfare and Ethics Review Committee of Yantai University.
Supplemental Material
Download JPEG Image (194.5 KB)Acknowledgements
Geolocation information: The location of sample collection was Jingouzhai, Yantai City, Shandong Province, China (37.5311 N, 121.4378 E).
Disclosure statement
The authors report there are no competing interests to declare.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. NC_046935. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA781747, SRR16979199, and SAMN21370456, respectively.
Additional information
Funding
References
- Boore JL, Brown WM. 1994. Complete DNA sequence of the mitochondrial genome of the black chiton, Katharina tunicata. Genetics. 138(2):423–443.
- Cui YT, Guo XY, Wang SS, Xu YR, Xiaoyue S, Li RR, Wang YH, Qu JY, Wang XM, Liu XM. 2019. The complete mitochondrial genome and phylogenetic analysis of Ischnochiton hakodaensis (Carpenter, 1893). Mitochondrial DNA B Resour. 4(2):2619–2621.
- Garcia-Ibanez S, Flores-Garza R, Flores-Rodriguez P, Violante-Gonzalez J, Valdes-Gonzalez A, Olea-de la Cruz FG. 2013. Fisheries diagnostic of Chiton articulatus (Mollusca: Polyplacophora) at Acapulco, Mexico. Rev Biol Mar Oceanogr. 48(2):293–302.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(1):59–64.
- Guerra D, Bouvet K, Breton S. 2018. Mitochondrial gene order evolution in Mollusca: inference of the ancestral state from the mtDNA of Chaetopleura apiculata (Polyplacophora, Chaetopleuridae). Mol Phylogenet Evol. 120:233–239.
- Guo XY, Cui YT, Wang SS, Xu YR, Sun XY, Li RR, Wang YH, Qu JY, Wang XM, Liu XM. 2019. The complete mitochondrial genome and phylogenetic analysis of Acanthochitona rubrolineatus (Lischke, 1873). Mitochondrial DNA B Resour. 4(2):2622–2624.
- Higo S, Callomon P, Goto Y. 1999. Catalogue and bibliography of the marine shell-bearing Mollusca of Japan. Osaka, Japan: Elle Scientific Publications, p. 749.
- Irisarri I, Uribe JE, Eernisse DJ, Zardoya R. 2020. A mitogenomic phylogeny of chitons (Mollusca: Polyplacophora). BMC Evol Biol. 20(1):22–36.
- Kirschvink JL, Lowenstam HA. 1979. Mineralization and magnetization of chiton teeth: paleomagnetic, sedimentologic, and biologic implications of organic magnetite. Earth Planet Sci Lett. 44(2):193–204.
- Owada M. 2018. Phylogenetic relationships among Japanese species of the genus Ischnochiton (Polyplacophora: Ischnochitonidae), including a new species. Zoolog Sci. 35(3):281–291.
- Pu DQ, Liu HL, Gong YY, Ji PC, Li YJ, Mou FS, Wei SJ. 2017. Mitochondrial genomes of the hoverflies Episyrphus balteatus and Eupeodes corollae (Diptera: Syrphidae), with a phylogenetic analysis of Muscomorpha. Sci Rep. 7:44300.
- Pu SW, Lu F, Huang B. 2020. The complete mitochondrial genome of Haliotis ovina. Mitochondrial DNA B. 5(3):2312–2313.
- Tamura K, Stecher G, Kumar S. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 38(7):3022–3027.
- Veale AJ, Williams L, Tsai P, Thakur V, Lavery S. 2016. The complete mitochondrial genomes of two chiton species (Sypharochiton pelliserpentis and Sypharochiton sinclairi) obtained using Illumina next generation sequencing. Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):537–538.
- Xu HW, Chu ZL, Zhang J, Jing MD, Huang L. 2020. Genetic diversity and population structure of Acanthochiton rubrolineatus (Polyplacophora) based on mitochondrial and nuclear gene markers. Diversity. 12(4):159–172.