Abstract
Japanese butterflyfish (Chaetodon nippon) belong to the family Chaetodontidae and order Chaetodontiformes. It has circular mitochondrial genome of 16,507 bp in length with 55.4% of A + T content and has 37 genes, including 22 tRNA, 2 rRNA, and 13 protein-coding genes, in addition to a control region. The results of phylogenetic analysis indicated that the C. nippon, C. wiebeli, C. auripes, C. auriga, C. octofasciatus, C. speculum, and C. modestus are closely related to each other. The findings of this study will provide useful genetic information for further phylogenetic and taxonomic classifications of Chaetodontidae.
Introduction
Japanese butterflyfish (Chaetodon nippon, Steindachner and Doederlein, 1883) belongs to the family Chaetodontidae and order Chaetodontiformes and can be found on shallow rocky reefs, off the shores of the Philippines, South Korea, Taiwan, and Japan. Fishes belonging to the family Chaetodontidae serve as an indicator fish group for the coral reef environment. The fish species in the genus Chaetodon are widely distributed; however, they are closely related based on morphology and selected mitochondrial genes (Hsu et al. Citation2007; Ferdyan et al. Citation2021). Mitogenomes have been widely used as molecular markers for evolutionary phylogeny and population genetics studies in a broad range of species. While no previous research on the complete mitogenome of C. nippon has been reported, we report the complete mitochondrial genome sequencing of C. nippon and its phylogenetic relationship with other Chaetodon species. The mitogenome sequence of C. nippon reported in this study will provide useful genetic data for species identification; the understanding of taxonomic classification, phylogenetic relationship, and the evolutionary history of the Chaetodontidae.
Materials
The C. nippon specimen was donated by Professor Hiroyuki Motomura, Kagoshima University Museum, Japan. It was collected from the coast of Uchinoura Bay in Japan (31°29′03.59″N 131°07′89.57″E) (). The specimen was deposited under the voucher number KAUM-131676 at the Marine Fish Resources Bank of Korea (MFRBK), Pukyong National University (PKNU), Busan, South Korea (Jin-Koo Kim, [email protected]).
Methods
Total genomic DNA was extracted from the muscle using the DNeasy Blood and Tissue Kit (Qiagen, Germany) according to the manufacturer’s instructions. The TrueSeq Nano DNA Kit was used to construct the DNA library, which was then sequenced with 150 bp paired-end reads on the Illumina platform (Illumina, HiSeq 2500, San Diego, CA, USA). The obtained reads were cleaned using cutadapt 1.9 (Martin Citation2011), and the low-quality reads (Q < 20) were removed. As shown in Figure S1, using SPAdes v3.13.0, the cleaned sequences were used for de novo assembly (Bankevich et al. Citation2012). Protein-coding, rRNA and tRNA genes were annotated using MitoFish (http://mitofish.aori.u-tokyo.ac.jp/) (Iwasaki et al. Citation2013)
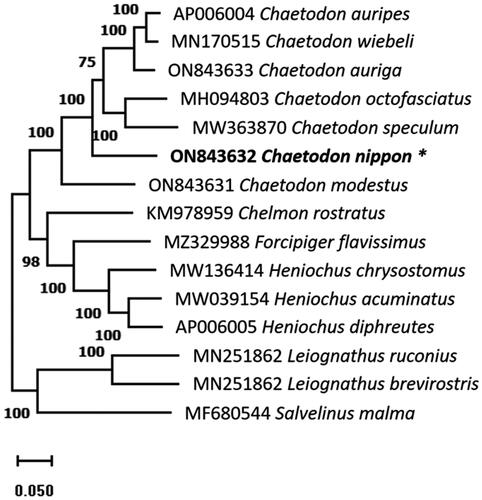
Complete mitochondrial genome sequences of C. nippon, 11 additional species from the family Chaetodontidae found in GenBank, and three species as outgroup members (Leiognathus brevirostris and L. ruconius from Leiognathidae and Salvelinus malma from Salmonidae) were used to construct a phylogenetic tree (). Prior to analysis, several mitochondrial genome sequences were aligned using ClustalW. Using MEGA11 v11.0.8 (Tamura et al. Citation2021) and MrBayes v3.2.6 (Huelsenbeck and Ronquist Citation2001), the phylogenetic tree was constructed using maximum likelihood (ML) with 1000 bootstrap.
Table 1. A list of the GenBank accession numbers of the complete mitochondrial genome sequences used in this study.
Results
The annotated complete mitochondrial genome sequence of C. nippon was submitted to NCBI (GenBank: ON843632). It was 16,507 bp in length, containing 37 genes, including 13 protein-coding, 2 rRNA and 22 tRNA genes, and a control region (D-loop, ). The overall base composition was 27.5%, 27.9%, 16.8%, and 27.8% for A, T, G, and C, respectively, with a slight A + T bias (55.4%). The 12S and 16S rRNA genes of C. nippon were positioned between the tRNA-Phe and tRNA-Leu genes, separated by the tRNA-Val gene. Additionally, several protein-coding genes, including ATP6, COX2, COX3, CYTB, ND2, and ND4, had incomplete stop codons. The ML phylogenetic trees constructed using MEGA11 () and MrBayes (Figure S2) were based on the complete mitochondrial genome sequences, showed that C. nippon is monophyletic with C. wiebeli, C. auripes, C. auriga, C. octofasciatus, C. speculum, and C. modestus.
Figure 2. Gene map of the mitochondrial genome of Japanese butterflyfish (Chaetodon nippon). Genes outside the circle are transcribed clockwise, whereas those inside are transcribed anticlockwise. Using MitoFish (http://mitofish.aori.u-tokyo.ac.jp/), a circular map was created by applying annotation results. Regions of protein-coding genes (PCGs) marked in black, tRNA in red, rRNA in yellow, and control region (D-loop) in brown.
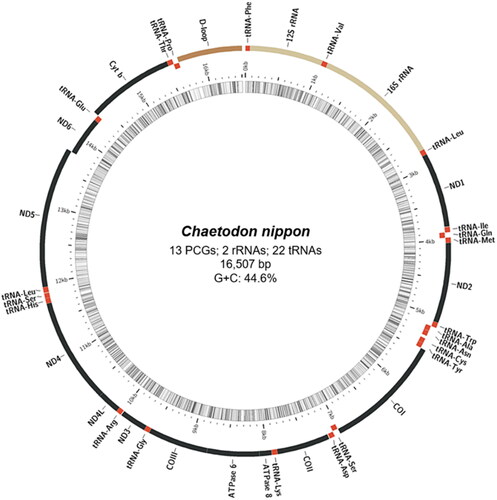
Figure 3. The phylogenetic tree was constructed using whole mitochondrial genome sequences of 15 species based on the maximum likelihood with 1000 bootstrap replicates using MEGA11. The bootstrap support values are shown by the numbers on the branches, and the species in this study is indicated by an asterisk next to its name.
Discussion and conclusion
The characteristic features of the C. nippon mitochondrial genome, such as A + T content, positions of 12S and 16S rRNA, and incomplete stop codons of protein-coding genes, are in good accordance with those of other Chaetodontidae species (Zhu et al. Citation2018; Yukai et al. Citation2019; Patil et al. Citation2022a, 2022b). The lengths of the mitochondrial genome and control region vary among Chaetodon species (Table S1). The difference in length has become a benchmark for research on interspecific genetic diversity, which may result in phylogenetic reconstructions of numerous closely related species. The phylogenetic tree shows that Chaetodon species are monophyletic and closely related, which is consistent with previous studies (Patil et al. Citation2022a, Citation2022b). The findings of this study may help future population genetics and evolution studies.
.
Ethical approval
The sample used for this study was a dead body of fish and as per the animal experimental ethics in the Republic of Korea (Standard operating guideline; IACUC – Institutional Animal Care and Use Committee, Book no. 11-1543061-000457-01, effective from Dec. 2020) does not need any approval from an Ethics Committee.
Authors’ contributions
M.P.P. and J-O.K. performed the experiments, analyzed the data, were involved in certain tools for the analysis, and drafting of the paper, and approved the final draft. Y.B.S., Y-J.L., and J-K.K. were involved in certain tools for analysis, sorting out the results, and preparing figures. G-D.K., involved in the conception and design of the work, funding acquisition, revising it critically for intellectual content, and the final approval of the version to be published.
Supplemental Material
Download MS Word (541.3 KB)Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/nuccore/ON843632) under accession no. ON843632. The associated BioProject, BioSample, and SRA numbers are PRJNA854332, SAMN29431623, and SRR19905897, respectively
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Ferdyan R, Razak A, Ahda Y. 2021. Comparison of the Chaetodontidae mitochondrial genome. Biota: biologi Dan Pendidikan Biologi. 14(1):46–55.
- Hsu KC, Chen JP, Shao KT. 2007. Molecular phylogeny of Chaetodon (Teleostei: Chaetodontidae) in the Indo-West Pacific: evolution in geminate species pairs and species groups. Raffles Bull Zool. 14:77–86.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30(11):2531–2540.
- Jiang F, Yang N, Huang H. 2022. Characterization and phylogenetic analysis of the mitochondrial genome sequence of Heniochus acuminatus. Mitochondrial DNA Part B Resour. 7(9):1694–1695.
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17(1):10–12.
- Patil MP, Kim J-O, Lee Y-J, Seo YB, Kim J-K, Kim G-D. 2022a. The complete mitochondrial genome of threadfin butterflyfish, Chaetodon auriga (Chaetodontiformes: chaetodontidae) and phylogenetic analysis. Mitochondrial DNA Part B Resour. 7(11):1922–1924.
- Patil MP, Kim J-O, Lee Y-J, Seo YB, Kim J-K, Kim G-D. 2022b. Complete mitochondrial genome of brown-banded butterflyfish Chaetodon modestus (Chaetodontiformes, Chaetodontidae) and phylogenetic analysis. Mitochondrial DNA Part B Resour. 7(11):2012–2014.
- Sui Y, Qin B, Song X, Sheng W, Zhang B. 2019. Complete mitochondrial genome of the deep pugnose ponyfish Secutor ruconius (Perciformes: Leiognathidae) in the East China Sea. Mitochondrial DNA Part B Resour. 4(2):3563–3564.
- Tamura K, Stecher G, Kumar S. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol Bio Evol. 38(7):3022–3027.
- Wang LJ, You F, Wu ZH. 2016. Complete mitochondrial genome of copperband butterflyfish Chelmon rostratus (Teleostei, Perciformes, Chaetodontidae). Mitochondrial DNA Part A DNA Mapp Seq Anal. 27(3):2141–2142.
- Yamanoue Y, Miya M, Matsuura K, Yagishita N, Mabuchi K, Sakai H, Katoh M, Nishida M. 2007. Phylogenetic position of tetraodontiform fishes within the higher teleosts: Bayesian inferences based on 44 whole mitochondrial genome sequences. Mol Phylogenet Evol. 45(1):89–101.
- Yang Y, Li T, Lin H, Huang X, Yu W, Huang Z. 2021. The complete mitochondrial genome of Chaetodon speculum (Chaetodontiformes, Chaetodontidae). Mitochondrial DNA Part B Resour. 6(4):1290–1291.
- Yang LG, Meng FX, Wang RX, Shi G. 2017. Complete mitochondrial genome of the Salvelinus malma sp. (Salmoniformes, Salmonidae) with phylogenetic consideration. Mitochondrial DNA Part B Resour. 2(2):889–890.
- Yu W, Yang Y, Qi Z, Shan B, Huang X, Liu Y, Lin H, Li T, Huang Z, Ma Z, et al. 2021. The complete mitochondrial genome of Heniochus chrysostomus (Perciformes, Chaetodontidae). Mitochondrial DNA Part B Resour. 6(3):933–935.
- Yukai Y, Xiaolin H, Heizhao L, Tao L, Wei Y, Zhong H. 2019. The complete mitochondrial genome of Chaetodon wiebeli (Chaetodontiformes, Chaetodontidae). Mitochondrial DNA Part B Resour. 4(2):3145–3146.
- Zhu K, Gong L, Lü Z, Liu L, Jiang L, Liu B. 2018. The complete mitochondrial genome of Chaetodon octofasciatus (Perciformes: Chaetodontidae) and phylogenetic studies of Percoidea. Mitochondrial DNA Part B Resour. 3(2):531–532.

