Abstract
The tree shrew (Tupaia belangeri) is currently placed in the order Scandentia. Owing to their unique characteristics, such as small body size, high brain-to-body mass ratio, short reproductive cycle and life span, and low maintenance costs in laboratory conditions, tree shrews have been proposed as alternative experimental animals to primates in biomedical research. In this study, we determined the complete mitochondrial genome of the subspecies Tupaia belangeri yaoshanensis (T.b. yaoshanensis). The mitochondrial DNA (mtDNA) is 16,777 bp long and contains 13 protein-coding genes (PCGs), two ribosomal RNA genes (12S and 16S), and 22 transfer RNA (tRNA) genes. The base composition of the mitogenome was A (32.28%), T (26.82%), G (14.79%), and C (26.11%). For the 13 PCGs, 1405 variable sites were found between T.b. yaoshanensis and T.b. chinensis (JN800724), of which 916 were synonymous and 489 were nonsynonymous. The frequency of mutations significantly varied among the different genes, with the highest value in the mt-NAD5 gene of tree shrews. Phylogenetic analysis based on the amino acid sequences of 13 PCGs revealed a closer relationship between the species of Scandentia and Lagomorpha. To the best of our knowledge, this is the first study to report the complete mitochondrial genome sequence of T.b. yaoshanensis.
Introduction
The tree shrew (Tupaia belangeri) is currently placed in the order Scandentia and is found throughout Southeast Asia and Southwest China (Wang Citation1987; Helgen Citation2005; Xu et al. Citation2012). This species exhibits unique characteristics that render it a potentially advantageous animal model, including a small body size, short reproductive cycle and life span, ease of handling, low maintenance costs in laboratory conditions, and genetic closeness to primates (Yao Citation2017). Wang (Citation1987) suggested that Chinese tree shrews could be classified into six subspecies according to their geographical, morphological, and morphometric data (Tupaia belangeri chinensis, Tupaia belangeri gaoligongensis, Tupaia belangeri modesta, Tupaia belangeri tonquinia, Tupaia belangeri yunalis, and Tupaia belangeri yaoshanensis), marking the first attempt at subspecies classification for Tupaia belangeri. Tupaia belangeri yaoshanensis (hereafter, T.b. yaoshanensis) is a unique tree shrew subspecies inhabiting the Dayao Mountains of Guangxi, China (106°38′E, 24°63′N), in the northwestern part of the province. It has the largest body shape among tree shrews in China, with a weight of approximately 180 g and body length of approximately 200 mm (). Previous studies have conducted mitochondrial genome sequencing of other tree shrew subspecies (accession nos.: JN800722–JN800724, NC_002521, Xu et al. Citation2012), but that of T.b. yaoshanensis has not yet been reported. We determined the complete mitochondrial genome of T.b. yaoshanensis to obtain more information regarding its phylogenetic status.
Materials
The specimen was provided by the Experimental Center of Artificial Domestication and Cultivation of Guangxi University of Chinese Medicine. The specimen and DNA were stored at Guangxi University of Chinese Medicine, under voucher no. YS062-02 (Dr. Haibo Tang, [email protected]).
Methods
Total genomic DNA from the liver tissue sample of T.b. yaoshanensis (no. YS062-02) was extracted using the phenol–chloroform extraction method (Green and Sambrook Citation2012). Based on the whole mitochondrial genome sequence of T.b. chinensis published in NCBI (accession no. JN800724, Xu et al. Citation2012), we used NCBI Primer-BLAST to design 14 pairs of primers that cover the whole mitochondrial DNA (mtDNA) (Table S1). The PCR products were purified on spin columns and sequenced using an ABI3730 Sequencing System (Sangon Biotech, Shanghai, China). The 14 sequences were spliced into a complete mitochondrial genome using DNAStar with the ‘Seqman’ function and finally submitted to GenBank (accession no. OP210313). The complete mitochondrial genome was annotated using the MITOS server (Bernt et al. Citation2013). Amino acid sequence homology alignment was performed using DNAStar with the ‘MegAlign’ function. Maximum-likelihood (ML) phylogeny was built using MEGA 7 with 1000 bootstrap replicates (Kumar et al. Citation2016; Nam et al. Citation2021).
Results
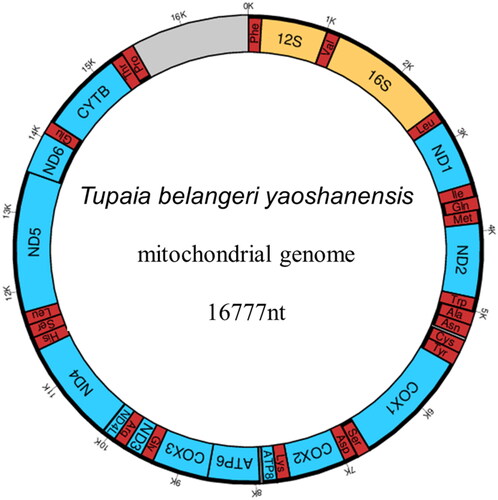
The total length of the T.b. yaoshanensis mitochondrial genome was 16,777 bp with a GC content of 40.9% (). The nucleotide composition was 32.28% A, 26.82% T, 14.79% G, and 26.11% C. The gene composition of the T.b. yaoshanensis mitogenome was 13 protein-coding genes (PCGs), two ribosomal RNA genes (12S and 16S), and 22 transfer RNA (tRNA) genes. For the 13 PCGs, a total of 1405 variable sites were found between T.b. yaoshanensis and T.b. chinensis (JN800724, Xu et al. Citation2012), of which 916 were synonymous and 489 were nonsynonymous. For amino acid sequence homology analysis, the 13 PCGs of tree shrews (T.b. yaoshanensis, T.b. chinensis) were 74.8–75% homologous to human PCGs, which is higher than those of mice (71.9%) and rabbits (74.6%). The mutation frequency varied significantly among the different genes, with the highest value observed in the mt-NAD5 gene of tree shrews. For amino acid sequence homology analysis, the mt-NAD5 of tree shrew (T.b. yaoshanensis, T.b. chinensis) was 68.7–69.7% homologous to human mt-NAD5, which is higher than that of mouse (65%) and rabbit (67.9%). Furthermore, T.b. yaoshanensis mt-NAD5 had the highest amino acid sequence identity with humans (69.7%).
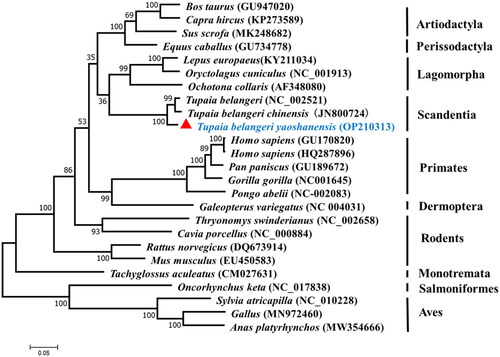
Phylogenetic analysis was inferred from the available mtDNA of T.b. yaoshanensis, other tree shrew subspecies, primates, rodents, lagomorpha, artiodactyla, perissodactyla, dermoptera, monotremata, salmoniformes, and aves based on the amino acid sequences of 13 PCGs (for accession numbers, see ). Phylogenetic analysis showed that T.b. yaoshanensis was clustered with T. belangeri and T.b. chinensis that were closely related to Lagomorpha, whereas Artiodactyla and Perissodactyla diverged after the Scandentia/Lagomorpha cluster. They formed a large evolutionary clade with Primates.
Discussion and conclusions
This study reports the mitogenome sequence of T.b. yaoshanensis for the first time, thus providing fundamental data for further exploration of mtDNA evolution in tree shrews.
Ethical approval
The experiment was reviewed and approved by Guangxi University of Chinese Medicine Institutional Animal Ethical and Welfare Committee (DW20201206-123).
Author contributions
Study conception and design: Haibo Tang and Jing Leng; data collection: Yingying Cao; analysis and result interpretation: Shanshan Zhai, Zhengtuan Huang, and Zhuxin Li; draft manuscript preparation: Yingying Cao, Junyu Tao, and Liang Liang; critical content revision of the manuscript: Yingying Cao and Jian Xiao.
Supplemental Material
Download MS Word (17.6 KB)Acknowledgements
We are grateful to Associate Professor Shaoqiang Ruan at the College of Foreign Languages, Guangxi University, for her help in editing the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are available in GenBank (https://www.ncbi.nlm.nih.gov/) under accession number OP210313. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA870698, SRR21118736, and SAMN30383116, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Green MR, Sambrook J. 2012. Molecular cloning: a laboratory manual. 4th ed. New York: Cold Spring Harbor Laboratory Press.
- Helgen KM. 2005. Order Scandentia. In: Wilson DE, Reeder DM, editors. Mammal species of the world: a taxonomic and geographic reference. 3rd ed. Maryland: Johns Hopkins University Press; p. 104–109.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Nam SE, Eom HJ, Park HS, Rhee JS. 2021. The complete mitochondrial genome of Lamprologus signatus (Perciformes: Cichlidae). Mitochondrial DNA B Resour. 6(12):3487–3489.
- Wang YX. 1987. Taxonomic research on Burma-Chinese tree shrew, Tupaia belangeri (Wagner), from Southern China. Zool Res. 8:213–230.
- Xu L, Chen SY, Nie WH, Jiang XL, Yao YG. 2012. Evaluating the phylogenetic position of Chinese tree shrew (Tupaia belangeri chinensis) based on complete mitochondrial genome: implication for using tree shrew as an alternative experimental animal to primates in biomedical research. J Genet Genomics. 39(3):131–137.
- Yao YG. 2017. Creating animal models, why not use the Chinese tree shrew (Tupaia belangeri chinensis). Zool Res. 38(3):118–126.