Abstract
The first complete mitochondrial genome of the freshwater goby Rhinogobius davidi was determined by high-throughput sequencing. This genome was 16,627 bp in length and consisted of 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and 2 non-coding control regions. Phylogenetic analysis based on the amino acid sequences of 13 mitochondrial protein-coding genes from R. davidi and 23 relatives suggested that R. davidi had a close mitogenome relationship with Rhinogobius giurinus. This complete genome of R. davidi will provide basal molecular data for future studies on taxonomy, comparative genomics, and adaptive evolution in Rhinogobius.
Introduction
Rhinogobius davidi (Sauvage & Dabry de Thiersant, 1874) is a benthic species of landlocked goby distributed in the Oujiang, Qiantang, and Yangtze River drainages, China (). This fish feeds on invertebrates and appears to be limited to smaller streams in the upper reaches of these large river drainages. There are no records of the species occurring in hydrostatic environments such as lakes in these drainages. The breeding period begins when the water temperature is above 20 °C. Before breeding, the male fish nests, and then attracts the females to enter and lay eggs. The eggs adhere to the ventral surface of the stones, and the male protects the eggs alone until they hatch. Female fish can reproduce once a month, each spawning up to 100 or more (Li 2011). Distinguishing morphological features of this species include: first dorsal fin VI, second dorsal fin I, 9–10, gluteal fin I, 6–8, pectoral fin 14–15, ventral fin I, 5; longitudinal scales 30–32, transverse scales 11, dorsal fin anterior scales 0–2 (mainly 0); vertebrae number 11 + 17 = 28; no tube hole in the sensory tube of the anterior gill cover; cheek with a brown vertical stripe below the eye (Yang et al. Citation2008; Li Citation2011).
Figure 1. The specimen of Rhinogobius davidi from Cao’e River, Shaoxing, Zhejiang Province, China (Photo by Lin Song).

Rhinogobius davidi is of ornamental value, but to date, it is poorly studied and what research has been done is limited to morphology and ecology (Chen and Miller 1998; Li 2011). Due to various manmade influences, the ecological environment of the basin is seriously damaged and the number of fish declined significantly. Genetic information is crucial to the development of strategies for the identification and management of this species (Yang et al. Citation2020). Therefore, this study is aimed to provide a first report on the mitochondrial genome of R. davidi and analyze the phylogenetic relationships among the genus Rhinogobius.
Materials
The specimens were obtained from Cao’e River in Shangyu District, Shaoxing, Zhejiang Province of China (29°57′17.94″N, 120°52′06.98″E), and were identified as R. davidi based on the morphological characters. Some specimens were kept in dry ice until DNA analysis and transferred to Shanghai Genesky Biotechnologies Inc, while others (Voucher number ASTIH-21b1108d23) were fixed in 95% ethanol and deposited in Aquatic Science and Technology Institution Herbarium (https://www.jsahvc.edu.cn/) with Lin Song ([email protected]) in charge. All animal handling and experimental procedures were performed in accordance with the recommendations of the Ethics Committee for Animal Experiments of Jiangsu Agri-animal Husbandry Vocational College (Taizhou, China).
Methods
The total genomic DNA was extracted from muscle tissue using the phenol-chloroform method (Barnett and Larson Citation2012). The DNA library was established with quality-controlled DNA samples, and amplified by high-fidelity polymerase to ensure sufficient library volume on the sequencer. Agilent 2100 Bioanalyzer (Agilent Technologies, USA) was applied to determine the size distribution of library fragments and evaluate the suitability for sequencing. After library pooling, next-generation sequencing was performed on Illumina HiSeq 4000 Sequencing platform (Illumina, CA, USA). The raw sequencing data were checked by FastQC, and removed of low-quality reads and adapter region with Trimmomatic (Bolger et al. Citation2014). The trimmed reads were mapped to the reference mitogenome of Rhinogobius giurinus (KU871066), using BWA v.0.7.17 (Li and Durbin Citation2009) with default parameters.
Samtool v.1.9 (Li and Durbin Citation2009) was employed to retrieve the aligned mitochondrial reads. The aligned reads with the mitogenome were then assembled using the software MetaSPAdes 3.13.0 (Nurk et al. Citation2017).
The resulting contig was annotated on MitoMaker 1.14 (Bernt et al. Citation2013). The final full mtDNA sequence is available in GenBank under accession number OM617724. And the genome circular map of R. davidi was drawn by CGView online server (https://proksee.ca/) (Grant and Stothard Citation2008).
We generated a phylogenetic tree with a set of all 22 Rhinogobius species available in GenBank and 2 Chaenogobius species. Each of the 13 PCGs was aligned separately using MUSCLE algorithm (Edgar Citation2004) in MEGA X, then the 13 PCG alignments were concatenated into a single multiple sequence alignment. The substitution model mtREV + G + I + F was selected utilizing the Find Best DNA/protein model tool of MEGA X software. The maximum likelihood analysis was performed on MEGA X with 1000 bootstrap replicates (Kumar et al. Citation2018). Chaenogobius gulosus (Oh et al. Citation2016) and Chaenogobius annularis were selected for the outgroups.
Results
The complete mitochondrial genome of R. davidi comprised 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (12S rRNA and 16S rRNA), and 2 non-coding control regions (control region and origin of light-strand replication), which formed a double-stranded circular molecule of 16,627 bp in length (). All other genes were encoded on the heavy strand (H-strand), except for one protein-coding gene (ND6) and eight tRNA genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer(UCN), tRNAGlu, and tRNAPro). The overall nucleotide composition in descending order was 17% for G, 25.9% for T, 27% for A, and 30.1% for C, with a slight AT-rich feature (52.9%).
Figure 2. Gene map of the mitochondrial genome of Rhinogobius davidi. Gene encoded on H- and L-strands with inverse arrow directions were shown outside and inside the circle, respectively. The complete mitogenome of R. davidi is 16,627 bp with the inclusion of 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, origin of L-strand replication (Ori-L) and control region (D-loop).
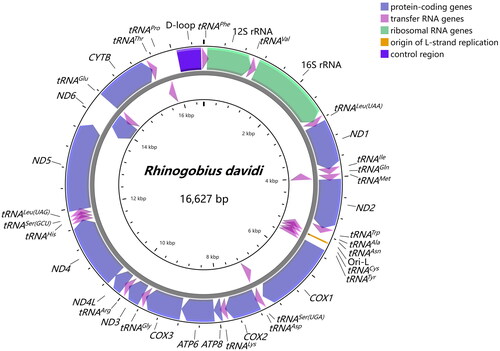
Twelve protein-coding genes began with the standard ATG, whereas COX1 initiated with GTG. There were both complete termination codons (TAA for ND1, COX1, ATP8, ATP6, ND4L, ND5; TAG for ND2, ND3, ND6) and incomplete termination codons (TA for COX3, T for COX2, ND4, CYTB). Four overlapping regions revealed between protein-coding genes were ATP8-ATP6, ATP6-COX3, ND4L-ND4, and ND5-ND6, and there were two overlaps between tRNAs (tRNAIle-tRNAGln, tRNAGln-tRNAMet). For rRNAs, 12S rRNA was located between tRNAPhe and tRNAVal with 954 bp in length, and 16S rRNA was located between tRNAVal and tRNALeu(UUR) with 1,661 bp in length. They were separated by tRNAVal with the same situation found in other gobies (Wang et al. Citation2019; Tan et al. Citation2020). 22 tRNAs ranged from 65 to 76 bp, which were conservative and scattered throughout the mitogenome of R. davidi. The control region, which was 476 bp long, was located between tRNAPhe and tRNAPro.
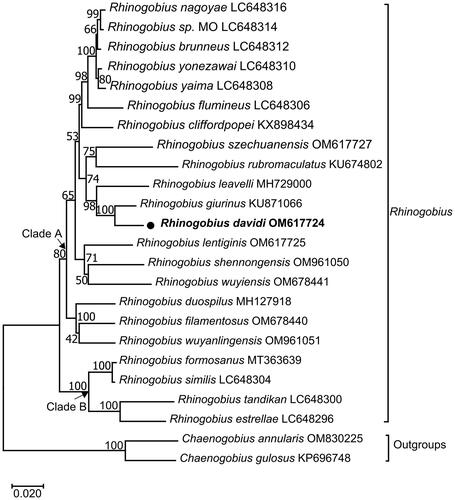
The maximum-likelihood phylogenetic tree showed the mitochondrial genome relationship of R. davidi in Rhinogobius. In , R. davidi clustered into a clade with R. giurinus (Xie et al. Citation2015), and then with R. leavelli (Zhang and Shen 2019). The three taxa, together with nine other Rhinogobius species (R. nagoyae (Maeda et al. Citation2021), R. sp. MO (Maeda et al. Citation2021), R. brunneus (Maeda et al. Citation2021), R. yonezawai (Maeda et al. Citation2021), R. yaima (Maeda et al. Citation2021), R. flumineus (Maeda et al. Citation2021), R. cliffordpopei (Wang et al. Citation2019), R. szechuanensis, R. rubromaculatus), constituted a sister-group to R. lentiginis, R. shennongensis and R. wuyiensis. All the above 15 Rhinogobius species formed Clade A with Rhinogobius duospilus (Tan et al. Citation2020), Rhinogobius filamentosus (Chen et al. Citation2022) and Rhinogobius wuyanlingensis (Song et al. Citation2022). In addition, Rhinogobius formosanus (Yang et al. Citation2020), Rhinogobius similis (Maeda et al. Citation2021), Rhinogobius estrellae (Maeda et al. Citation2021) and Rhinogobius tandikan (Maeda et al. Citation2021) formed Clade B, which was sister to Clade A.
Figure 3. Maximum-likelihood (ML) phylogenetic tree from amino acid sequences of 13 PCGs of Rhinogobius davidi and other 23 fishes. Accession numbers were indicated after the species names. The tree topology was evaluated by 1000 bootstrap replicates. Bootstrap values at the nodes correspond to the support values for ML methods. The tree was drawn to scale, with branch lengths measured in the number of substitutions per site.
Discussion and conclusion
We reported the first complete mitogenome sequencing of R. davidi by high-throughput sequencing and assembly. The arrangement and orientation of all 37 genes were in accordance with those goby mitogenomes published previously (Xie et al. Citation2015; Wang et al. Citation2019). In this study, more molecular information from mitogenomes of Rhinogobius was used to reconstruct the phylogenetic mitochondrial genome relationships, and the ML tree placed R. davidi in a well-supported cluster with Rhinogobius giurinus. The overall topology was similar to those of previous studies (Zhang and Shen 2019; Tan et al. Citation2020). The results presented here will be essential to the specimen identification, fishery resources management, and further phylogenetic studies on Gobionellinae species.
Ethical approval
Experiments were performed in accordance with the recommendations of the Ethics Committee for Animal Experiments of Jiangsu Agri-animal Husbandry Vocational College. These policies were enacted according to the Chinese Association for the Laboratory Animal Sciences and the Institutional Animal Care and Use Committee (IACUC) protocols.
Author contributions
LS and X-JC: conceptualization and workflow design; YS and QW: data acquisition and analysis. All authors were involved in drafting the paper and final version approval. The contributions are ranked in order.
Supplemental Material
Download (8.4 MB)Supplemental Material
Download MS Excel (18.5 KB)Supplemental Material
Download JPEG Image (788.1 KB)Supplemental Material
Download MS Excel (22.5 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the reference number OM617724. The associated “BioProject", “Bio-Sample” and “SRA” numbers are PRJNA808178, SAMN26030965, and SRR18064556 respectively.
Additional information
Funding
References
- Barnett R, Larson G. 2012. A phenol-chloroform protocol for extracting DNA from ancient samples. Methods Mol Biol. 840:13–19.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bolger A, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Chen IS, Miller PJ. 1998. Redescription of a Chinese freshwater goby, Gobius davidi (Gobiidae), and comparison with Rhinogobius lentiginis. Cybium. 22(3):211–221.
- Chen XJ, Song L, Liu WZ, Wang Q. 2022. Characteristic and phylogenetic analyses of mitochondrial genome for Rhinogobius filamentosus (Teleostei: Gobiidae: Gobionellinae), an endemic species in China. Mitochondrial DNA B Resour. 7(9):1752–1755.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797.
- Grant JR, Stothard P. 2008. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36(Web Server issue):W181–W184.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Luo F, Huang J, Luo T, Yang J, Wen Y. 2019. Complete mitochondrial genome and phylogenetic analysis of Yunnanilus pulcherrimus (Cypriniformes, Nemacheilidae). Mitochondrial DNA Part B. 4(1):1269–1270.
- Li F. 2011. Study on classification and distribution of Rhinogobius (Perciformes: Gobiidae) from the Qiantangjiang Basin. [Doctoral dissertation]. Shanghai: Fudan University. https://cdmd.cnki.com.cn/Article/CDMD-10246-1014444614.htm
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25(14):1754–1760.
- Maeda K, Shinzato C, Koyanagi R, Kunishima T, Kobayashi H, Satoh N, Palla HP. 2021. Two new species of Rhinogobius (Gobiiformes: oxudercidae) from Palawan, Philippines, with their phylogenetic placement. Zootaxa. 5068(1):81–98.
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. MetaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834.
- Oh J, Kim TW, Kim S. 2016. The complete mitochondrial genome of Chaenogobius gulosus (Gobiidae, Perciformes) from the South Sea, Korea. Mitochondrial DNA A DNA Mapp Seq Anal. 27(6):4207–4208.
- Song L, Chen XJ, Mao HX, Wang Q. 2022. Characterization and phylogenetic analysis of the complete mitochondrial genome of Rhinogobius wuyanlingensis (Gobiiformes: Gobiidae: Gobionellinae). Mitochondrial DNA B Resour. 7(7):1323–1325.
- Tan HY, Yang YY, Zhang M, Chen XL. 2020. The complete mitochondrial genome of Rhinogobius duospilus (Gobiidae: Gobionellinae). Mitochondrial DNA B Resour. 5(3):3406–3407.
- Wang D, Dai CX, Li Q, Li Y, Liu ZZ. 2019. Complete mitochondrial genome and phylogenic analysis of Rhinogobius cliffordpopei (Perciformes, Gobiidae). Mitochondrial DNA B Resour. 4(2):2473–2474.
- Xie LP, Yang XF, Ma ZH, Yang RB. 2015. Complete mitochondrial genome of Rhinogobius giurinus (Perciformes: Gobiidae: Gobionellinae). Mitochondrial DNA. 26(2):321–322.
- Yang JQ, Wu HL, Chen IS. 2008. A new species of Rhinogobius (Teleostei: Gobiidae) from the Feiyunjiang basin in Zhejiang Province, China. Ichthyological Research. 55:379–385.
- Yang CJ, Chen Y, Chen Z, He GH, Zhong ZL, Xue WB. 2020. The next-generation sequencing reveals the complete mitochondrial genome of Rhinogobius formosanus (Perciformes: Gobiidae). Mitochondrial DNA B Resour. 5(3):2673–2674.
- Zhang FB, Shen YJ. 2019. Characterization of the complete mitochondrial genome of Rhinogobius leavelli (Perciformes: Gobiidae: Gobionellinae) and its phylogenetic analysis for Gobionellinae. Biologia. 74:493–499.
