Abstract
The Ovis aries Duoma breed is one of the most important alpine grassland-type sheep breeds in China. In this current study, we first reported the complete mitochondrial genome of Duoma sheep. Mitochondrial genome is 16,618 bp in length and exhibits the same typical structure as the other published sheep breeds. The genome contains 37 genes (22 tRNA, two rRNA, and 13 protein-coding genes (PCGs)) and a control region (D-loop region). Phylogenetic analysis shows that the Duoma sheep breed is closer to Ganjia sheep. Our findings will help the further evolution and conservation studies of Duoma sheep.
Introduction
Duoma sheep (Ovis aries), also known as Anduo sheep, is one of the primitive Tibetan sheep breeds in China (). It is a short thin-tailed breed residing at elevations above 4850 m. Under high-altitude, UV radiation, hypoxia, and sparse vegetation conditions for a long time, Duoma sheep has evolved to be a prominent local sheep breed well-adapted to harsh environments in alpine cold pastoral areas. It is getting popular for herders to raise Duoma sheep due to the excellent properties of larger body size, higher wool output, high slaughter rate, and stronger environmental adaptation. Duoma is mainly white-coat coarse wool sheep breed with brown-white striped face, only a small proportion of sheep are brown coat. Their eyelids and lip are mainly in black, approximately 20% are brown and only a few are pink. Large curved horns are existed in vast majority male Duoma sheep, and female is scars or polled. To date, lots of sheep mitogenomes have already been published. However, limited information exists on Duoma mitogenome and its ancestral maternal sources. Here, we assembled and annotated the complete mitochondrial genome of Duoma sheep. We next performed a comparative genomic analysis among nine representative sheep breeds and the other two ovis species. The results herein provide valuable insights into the structure and constitute of Duoma mitogenome, and promote the protection and utilization of germplasm resources of Duoma sheep.
Figure 1. The reference image of Duoma sheep. Duoma sheep were white or brown coated with the black or brown or mixed color around the eyes and noses, their eyelids and lips are black or brown and occasional pink. Large curved horns are existed in vast majority male Duoma sheep, and female are mainly with small curved or polled. This reference image was taken by Tianzeng Song, the first author listed in this manuscript

Material
The blood specimen was collected from a living individual from a standard Duoma sheep population, the sampling location was in Duoma Township, Anduo County, Tibet Autonomous Region, China (32°1015′N, 91°6006′E) and stored in the State Key Laboratory of Sheep Genetic Improvement and Healthy Production of Xinjiang Province, China (contact person: Mengsi Xu; email address: [email protected]) under the voucher number 20200148.
Methods
Total DNA was extracted from the blood sample using a TIANamp Genomic DNA Kit (Tiangen, Beijing, China). Six pairs of primers were designed to amplified six 2–3-kb PCR products covering the whole mitogenome (Table S1; Figure S1). The mitogenome was successfully assembled using the sequencing data of six overlapping mtDNA amplified fragments (Gan et al. Citation2016). DNAStar 5.01 was used to assemble mitogenome (Kumar et al. Citation2016). The assembled Duoma mitogenome was then annotated with ContigExpress software using the Sheep mitogenome as the reference (GenBank accession: NC001941.1). Finally, the complete Duoma mitogenome sequence was deposited in GenBank under the accession number MW427606.1.
Results
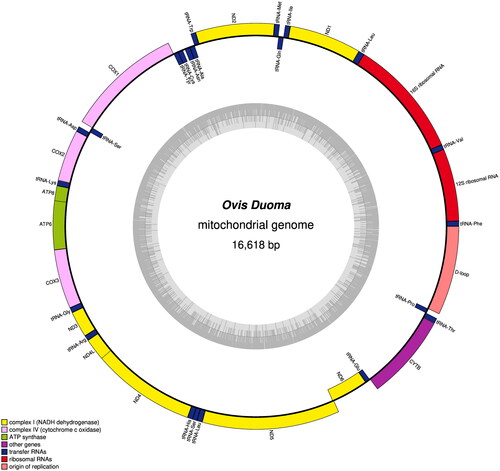
The complete mitochondrial genome of Duoma sheep is 16,618 bp (), including 22 tRNA genes, two rRNA genes, 13 protein-coding genes (PCGs), and a control region (D-loop), which structure and composition are similar to those of the other sheep breeds (Hiendleder et al. Citation1998; Fu et al. Citation2019; Qiao et al. Citation2020). Three PCGs (ND2, ND3, and ND5) used ATA as start codon and the other PCGs (ND1, COX1, COX2, COX3, ATP8, ATP6, ND4L, ND4, ND6, and Cyt b) were ATG. The 13 PCGs ended with five types of stop codons (TAA, TAG, AGA, TA, and T). Seven reading-frame overlaps were found in the Duoma mitochondrial genome: including ATP6 and ATP8 overlap by 40 nucleotides, ND6 and ND5 overlap by 17 nucleotides, ND4 and ND4L overlap by seven nucleotides, tRNASer and COX1 overlap by three nucleotides, tRNAGln and tRNAIle overlap by three nucleotides, COX3 and ATP6 overlap by one nucleotide, and tRNAPro and tRNAThr overlap by one nucleotide. The length of the 22 tRNAs genes ranges between 60 bp (tRNA-Ser) and 75 bp (tRNA-Leu).
Figure 2. The genome map of Duoma sheep mitogenome. Circle plot was colored according to gene functional categories. The complete mitogenome of Duoma sheep is 16,618 bp, including 22 tRNA genes, two rRNA genes, 13 protein-coding genes (PCGs), and a control region (D-loop).
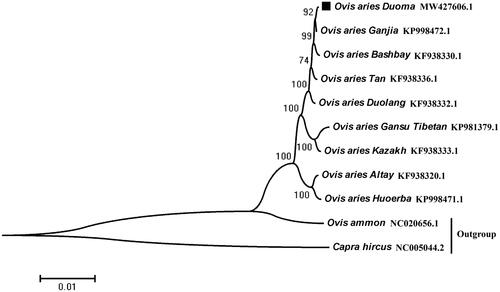
To evaluate the phylogenetic relationship of Duoma sheep with other sheep breeds, we downloaded the mitogenomes of eight sheep breeds from the GenBank (Lv et al. Citation2015), these eight mitogenomes were compared with that of the Duoma breed, and Ovis ammon and Capra hircus were used as the outgroup species. The maximum-likelihood phylogenetic tree was constructed using MEGA7.0 with 1000 bootstrap replicates. According to phylogenetic tree analysis, Duoma sheep has the closest genetic relationship with Ganjia sheep, and further genetic distance with Huoerba ().
Discussion and conclusions
The physical–geographical environmental conditions in Tibet are diverse and complex. The Qinghai-Tibetan Plateau encompasses an enormous range of altitude and latitude, which creates species and genetic diversity. All sheep in this region are known as Tibetan sheep. They are adapted to the long and cold winters and a poor diet, even thriving under such conditions (Cui et al. Citation2019; Wang et al. Citation2019; Liu et al. Citation2020). Their body size and production performance were significantly different, which were affected by environmental condition and artificial selection. Meanwhile, their mitochondrial genomes were quite distinct. A study by Li et al. found that the Tibetan sheep achieved the divergence and evolved into different breeds in response to adaptation to different geographic conditions, especially altitude, about 2500 years ago in the Qinghai Plateau region (Li et al. Citation2021). In the current research, four Tibetan sheep breeds were included in the phylogenetic tree analysis, with Duoma sheep being closely related to Ganjia sheep but being further genetically distanced from the other two Tibetan sheep breeds (Gansu Tibetan and Huoerba), this apparent discrepancy which may reflect the different altitudes and ecological environments of the home ranges of the different breeds of Tibetan sheep, and may also be affected by provenance hybridization during breed formation. This study of Duoma mitochondrial genome will provide more genomic resources for the Duoma sheep identification.
Ethical approval
Procedures were performed according to the Guide for the Care and Use of Laboratory Animals and approved by the Experimental Animal Care and Use Committee of Xinjiang Academy of Agricultural and Reclamation Sciences (Shihezi, China, ethic committee approval number: XJNKKXY-AEP-024, 18 April 2022).
Author contributions
Conceptualization, Ping Zhou; formal analysis, Mengsi Xu and Tianzeng Song; data analysis, Peng Yang and Mengjun Liu; methodology, Dawa Qiongda and Ciren Pubu; data interpretation, Chilie Baisang and Zha Xi; data acquisition, Ciren Wangmu and Danzeng Nima; writing draft, Mengsi Xu and Tianzeng Song; review and editing, Ping Zhou.
Supplemental Material
Download MS Word (34.5 KB)Supplemental Material
Download JPEG Image (381.8 KB)Supplemental Material
Download MS Word (28 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MW427606.1.
Additional information
Funding
References
- Cui XX, Wang ZF, Yan TH, Chang SH, Wang H, Hou FJ. 2019. Rumen bacterial diversity of Tibetan sheep (Ovis aries) associated with different forage types on the Qinghai-Tibetan Plateau. Can J Microbiol. 65(12):859–869.
- Fu DH, Ma XM, Jia CJ, Lei QY, Wu XY, Chu M, Ding XZ, Bao PJ, Pei J, Guo X, et al. 2019. Characterization of the complete mitochondrial genome sequence of Jialuo sheep (Ovis aries). Mitochondrial DNA B Resour. 4(2):2116–2117.
- Gan S-Q, Xu M-S, Meng J-M, Tang H, Li J, Yuan W, Wang X, Wang L, Zhou P. 2016. The complete mitochondrial genome of Marco Polo wild sheep (Ovis ammon polii). Mitochondrial DNA B Resour. 1(1):92–93.
- Hiendleder S, Lewalski H, Wassmuth R, Janke A. 1998. The complete mitochondrial DNA sequence of the domestic sheep (Ovis aries) and comparison with the other major ovine haplotype. J Mol Evol. 47(4):441–448.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Li LL, Ma SK, Peng W, Fang YG, Duo HR, Fu HY, Jia GX. 2021. Genetic diversity and population structure of Tibetan sheep breeds determined by whole genome resequencing. Trop Anim Health Prod. 53(1):174–192.
- Liu H, Hu L, Han X, Zhao N, Xu T, Ma L, Wang X, Zhang X, Kang S, Zhao X, et al. 2020. Tibetan sheep adapt to plant phenology in alpine meadows by changing rumen microbial community structure and function. Front Microbiol. 11:587558–587574.
- Lv FH, Peng WF, Yang J, Zhao YX, Li WR, Liu MJ, Ma YH, Zhao QJ, Yang GL, Wang F, et al. 2015. Mitogenomic meta-analysis identifies two phases of migration in the history of eastern Eurasian sheep. Mol Biol Evol. 32(10):2515–2533.
- Qiao GY, Zhang H, Zhu SH, Yuan C, Zhao HC, Han M, Yue YJ, Yang BH. 2020. The complete mitochondrial genome sequence and phylogenetic analysis of Alpine Merino sheep (Ovis aries). Mitochondrial DNA B Resour. 5(1):990–991.
- Wang G, He Y, Luo Y. 2019. Expression of OPA1 and Mic60 genes and their association with mitochondrial cristae morphology in Tibetan sheep. Cell Tissue Res. 376(2):273–279.