Abstract
We report the complete mitochondrial genome of Rhinogobius lentiginis, which was found to be a circular molecule of 16,633 bp in length and included 13 protein-coding genes (PCGs), 2 rRNA genes, 22 tRNA genes, and a non-coding control region. The overall base composition was 28.44% A, 26.21% T, 16.33% G, and 29.02% C. Phylogenetic analyses using maximum-likelihood and Bayesian inference methods revealed a close genome relationship among R. lentiginis, R. niger, R. shennongensis and R. maculagenys. The complete mitogenome of R. lentiginis will provide a valuable resource for species classification and conservation.
Introduction
Rhinogobius lentiginis (Gobiiformes: Gobiidae: Gobionellinae), an endemic species in China, is mainly distributed in the Lingjiang Basin, the Tingpangxi Basin, lower reaches of the Yangtze Basin and the Cao’ejiang River, which is a tributary of the Qiantang River Basin. R. lentiginis is a benthic, land-locked species that prefers slow-flowing water environment with fine gravel as the sediment. Human activities, such as the discharge of domestic and industrial sewage, stream dredging operations, and the construction of dams and reservoirs, will have a great impact on the survival of the goby. Taking the Qiantang River basin as an example, it was reported that there were 11 species of goby in 2011, but several of them were extremely rare. The population of these rare species is likely to decline or even become extinct due to the continuous deterioration of the basin environment (Li Citation2011). To develop strategies for the management of fisheries resources, the taxonomic survey and research on stream fish will be the primary work. Mitochondrial DNA is considered to be one of the most efficient and reliable molecular marker for species identification, evolutionary biology, population genetics and conservation biology (Ko et al. Citation2018). Therefore, we sequenced the complete mitogenome of R. lentiginis to present basic genetic information.
Materials
A specimen of R. lentiginis () was captured by the fish traps from Lingjiang River, Linhai City, Zhejiang Province, China (28°50′52.06″N, 121°09′23.32″E), in April 2022, and was euthanized with tricaine methanesulfonate (MS-222, MilliporeSigma, St. Louis, MO, USA). The specimen was given voucher number ASTIH-21b1108d24 and preserved in 95% ethanol at the Aquatic Science and Technology Institution Herbarium (https://www.jsahvc.edu.cn/, Lin Song, [email protected]).
Figure 1. The specimen of Rhinogobius lentiginis from Lingjiang River, Linhai City, Zhejiang Province, China. The main identifiable morphological features are the first dorsal fin VI; the second dorsal fin I, 8; gluteal fin I, 7–8 (mainly 7); pectoral fin 14–15; longitudinal scales 30–32; transverse scales 10–11; dorsal fin anterior scale 0; vertebrae number 26–27 (mainly 27); complete type of sensory canal pores (Li Citation2011). (Photo by Lin Song).

Methods
Genomic DNA was isolated from muscle tissue using the Tguide Cell/tissue genomic DNA Extraction Kit (OSR-M401, Tiangen, Beijing, China), followed by quality control to ensure sample concentration, purity and integrity using the NanoDrop 2000 (Thermo Fisher Scientific, USA). A DNA library consisting of all sheared mitochondrial DNA was prepared using the MGIEasy DNA Library Prep Kit (MGI Technology, Shenzhen, China). The purificaiton and size selection of the library was completed using Agencourt SPRIselect (Beckman Coulter, USA). The Agilent 2100 Bioanalyzer (Agilent Technologies, USA) was used to check the quality of the quality of the library. Finally, the constructed library was sequenced on the Illumina HiSeq 4000 Sequencing platform (Illumina, CA, USA) with paired-end reads (150 bp).
The sequencing reads were assessed for quality using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc), and low-quality reads as well as adapter sequences were trimmed using Trimmomatic ver. 0.40 (Bolger et al. Citation2014). The clean reads were mapped to the reference mitogenome of Rhinogobius brunneus (KT601096) using BWA v.0.7.17 (Li and Durbin Citation2009) with default parameters. Samtool v.1.9 (Li and Durbin Citation2009) was employed to retrieve the aligned mitochondrial reads. Then the retrieved reads were assembled with MetaSPAdes 3.13.0 (Nurk et al. Citation2017). The complete mitochondrial genome annotation was performed in the MitoFish webserver (http://mitofish.aori.u-tokyo.ac.jp/, Iwasaki et al. Citation2013). The mitogenome map was drawn by Proksee (https://proksee.ca/), an updated version of the CGView web server (Grant and Stothard Citation2008).
All available 25 mitogenomes of Rhinogobius species in GenBank were retrieved to analyze the phylogenetic position of R. lentiginis in the genus (). Two Tridentiger species, belonging to the same Gobionellinae family, were used as an outgroup. The complete mitogenome sequences were aligned by ClustalW in MEGA X (Kumar et al. Citation2018). The best fit substitution model (GTR + I + G) was calculated by jModelTest 2.1.10 (Darriba et al. Citation2012) under the Akaike information criterion (AIC). The maximum likelihood (ML) phylogenetic tree was built with bootstrap of 1000 replications using MEGA X, and the Bayesian inference (BI) tree was reconstructed by MrBayes 3.2.6 (Ronquist et al. Citation2012) with two independent runs and four Markov Monte Carlo (MCMC) chains. Each run consisted of 2,000,000 generations, sampled every 100 generations with 25% of the initial trees discarded as burn-in.
Table 1. Species names, GenBank accession numbers, and references of all 25 mitogenomes used in phylogeny reconstruction.
Results
Mitogenome organization
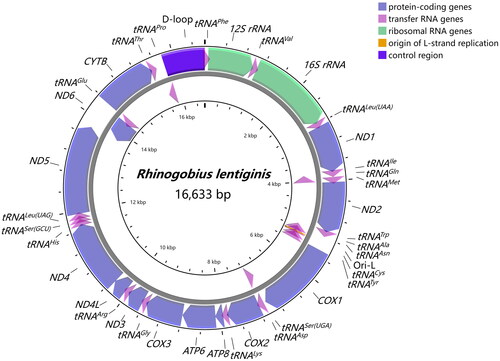
The complete mitogenome of R. lentiginis (GenBank accession number OM617725) was composed of 13 protein-coding genes (PCGs), 2 rRNA genes, 22 tRNA genes, and a non-coding control region (). The length of the mitogenome was 16,633 bp, with the overall base composition of 28.44% A, 26.21% T, 16.33% G, 29.02% C. Of all 37 mitochondrial genes, 28 were encoded on the heavy strand and 9 were encoded on the light strand (ND6, tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer(UCN), tRNAGlu, and tRNAPro). Most of the 13 PCGs contained the typical start codon ATG, except for COX1 starting with GTG, consistent with other fish mitochondrial genomes (Tan et al. Citation2020; Yang et al. Citation2020). Conventional stop codons were observed in ten PCGs (TAA for ND1, ND2, COX1, ATP8, ATP6, COX3, ND4L, ND5, and TAG for ND3, ND6), while the incomplete stop codon T was observed in the remaining PCGs (COX2, ND4, and CYTB). The lengths of the PCGs varied from 165 to 1,839 bp. The 12S rRNA (950 bp) and the 16S rRNA (1,688 bp) were separated by tRNAVal. Total 22 tRNAs ranged from 66 bp (tRNACys) to 76 bp (tRNALys) in length. The control region of 842 bp was identified between tRNAPro and tRNAPhe.
Phylogenetic analysis
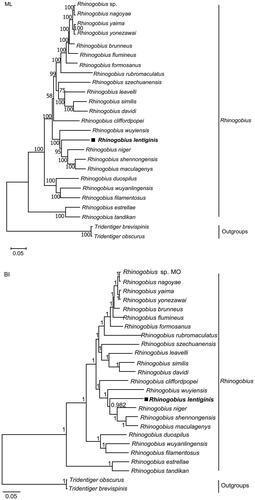
Both reconstruction methods recovered that there were two main clades in Rhinogobius (), in which R. estrellae and R. tandikan formed one, and other 21 Rhinogobius species formed the other clade. In our analyses, both trees displayed that R. lentiginis had a close mitochondrial genome relationship with R. niger, R. shennongensis and R. maculagenys. The genetic distance between R. lentiginis and 22 other Rhinogobius species ranged from 0.109 to 0.183.
Figure 3. Maximum-likelihood (a) and Bayesian inference (b) phylogenetic trees inferred from complete mitochondrial genomes of Rhinogobius lentiginis and other 24 fishes. Numbers at nodes represent bootstrap support values for ML tree, and posterior probabilities for BI tree. The discordant result between the two trees was the placement of R. cliffordpopei.
Discussion and conclusion
The complete mitogenome of the freshwater goby R. lentiginis had highly conserved structural organization and typical gene content, which were similar to Gobionellinae fishes (Wang et al. Citation2019; Yang et al. Citation2020). Our phylogenetic analyses of R. lentiginis revealed a close mitogenome relationship with R. niger, R. shennongensis and R. maculagenys. Present grouping results provided a better resolution for the genetic relationships among members of the Rhinogobius genus. Previous studies on the phylogeny of Rhinogobius species have shown that changes of members in the phylogenetic tree can affect the relationships of the species (Yamasaki et al. Citation2015; Wang et al. Citation2019). Therefore, further studies including morphology and genetics based on extensive taxon sampling are needed to assess the phylogenetic relationships among Rhinogobius species and genera. Overall, our results provide the genetic basis for resource conservation.
Ethical approval
Experiments were performed in accordance with the recommendations of the Ethics Committee for Animal Experiments of Jiangsu Agri-Animal Husbandry Vocational College. These policies were enacted according to the Chinese Association for the Laboratory Animal Sciences and the Institutional Animal Care and Use Committee (IACUC) protocols.
Author contributions
LS and X-J C: conceptualization and workflow design; Y-W G and QW: data acquisition and analysis. All authors were involved in drafting the paper and final version approval. The contributions are ranked in order.
Supplemental Material
Download MS Excel (18.5 KB)Supplemental Material
Download MS Excel (22.5 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the reference number OM617725. The associated ‘BioProject,’ ‘Bio-Sample’ and ‘SRA’ numbers are PRJNA808181, SAMN26031220, and SRR18131289 respectively.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Chen TT, Ren MD, Li QQ, Xie QM, Su SP, Li XL. 2019. Characterization and phylogenetic analysis of the complete mitochondrial genome of Rhinogobius sp. (Perciformes, Gobiidae). Mitochondrial DNA Part B. 4(2):3126–3127.
- Chen XJ, Song L, Liu WZ, Wang Q. 2022. Characteristic and phylogenetic analyses of mitochondrial genome for Rhinogobius filamentosus (Teleostei: Gobiidae: Gobionellinae), an endemic species in China. Mitochondrial DNA Part B. 7(9):1752–1755.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772–772.
- Gong L, Du X, Lü ZM, Liu LQ. 2017. The complete mitochondrial genome characterization of Tridentiger obscurus (Gobiiformes: Gobiidae) and phylogenetic analyses of Gobionellinae. Mitochondr DNA Part B. 2(2):662–663.
- Grant JR, Stothard P. 2008. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36(Web Server):W181–W184.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30(11):2531–2540.
- Ko AM, Zhang Y, Yang MA, Hu Y, Cao P, Feng X, Zhang L, Wei F, Fu Q. 2018. Mitochondrial genome of a 22,000-year-old giant panda from southern China reveals a new panda lineage. Currr Biol. 28: R693–R694.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Li F. 2011. Study on classification and distribution of Rhinogobius (Perciformes: Gobiidae) from the Qiantangjiang Basin [Doctoral dissertation]. Shanghai: Fudan University. http://cdmd.cnki.com.cn/Article/CDMD-10246-1014444614.htm
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25(14):1754–1760.
- Liu WZ, Song L, Chen XJ, Liu HX. 2023. The complete mitochondrial genome of Rhinogobius szechuanensis (Gobiiformes: Gobiidae: Gobionellinae). Mitochondrial DNA Part B. 8(2):192–196.
- Maeda K, Shinzato C, Koyanagi R, Kunishima T, Kobayashi H, Satoh N, Palla HP. 2021. Two new species of Rhinogobius (Gobiiformes: Oxudercidae) from Palawan, Philippines, with their phylogenetic placement. Zootaxa. 5068(1):81–98.
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. MetaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systemat Biol. 61(3):539–542.
- Shang YQ, Wang XB, Liu G, Wu XY, Wei QG, Sun GL, Mei XS, Dong YH, Sha WL, Zhang HH. 2022. Adaptability and evolution of Gobiidae: a genetic exploration. Animals. 12(14):1741.
- Song L, Chen XJ, Mao HX, Wang Q. 2022. Characterization and phylogenetic analysis of the complete mitochondrial genome of Rhinogobius wuyanlingensis (Gobiiformes: Gobiidae: Gobionellinae). Mitochondr DNA Part B. 7(7):1323–1325.
- Tan HY, Yang YY, Zhang M, Chen XL. 2020. The complete mitochondrial genome of Rhinogobius duospilus (Gobiidae: Gobionellinae). Mitochondr DNA Part B. 5(3):3406–3407.
- Wang D, Dai CX, Li Q, Li Y, Liu ZZ. 2019. Complete mitochondrial genome and phylogenic analysis of Rhinogobius cliffordpopei (Perciformes, Gobiidae). Mitochondrial DNA Part B. 4(2):2473–2474.
- Yang CJ, Chen Y, Chen Z, He GH, Zhong ZL, Xue WB. 2020. The next-generation sequencing reveals the complete mitochondrial genome of Rhinogobius formosanus (Perciformes: Gobiidae). Mitochondr DNA Part B. 5(3):2673–2674.
- Yamasaki YY, Nishida M, Suzuki T, Mukai T, Watanabe K. 2015. Phylogeny, hybridization, and life history evolution of Rhinogobius gobies in Japan, inferred from multiple nuclear gene sequences. Mol Phylogenet Evol. 90:20–33.
- Zhang FB, Shen YJ. 2019. Characterization of the complete mitochondrial genome of Rhinogobius leavelli (Perciformes: Gobiidae: Gobionellinae) and its phylogenetic analysis for Gobionellinae. Biologia. 74(5):493–499.