Abstract
Bidens pilosa L. 1753 is a perennial herbaceous flowering plant, traditionally used in foods and medicines. In this study, we sequenced, assembled, and characterized the complete plastome of B. pilosa from Beijing, China. The plastome (MN385242) is circularized with a conservative quadripartite structure. Its length is 150,524 bp, including a large single-copy region (83,535 bp), a small single-copy region (17,627 bp), and a pair of inverted repeat regions (each 24,681 bp). The plastome consists of 128 genes, including 78 unique protein-coding, 28 unique tRNA, and 4 unique rRNA genes. Phylogenetic analyses showed all five B. pilosa plants couldn’t form a monophyletic clade and were separated into three clades. The results of K2P distance and molecular markers were all consistent with those of phylogenetic analysis, revealing high genetic diversity and even possible misidentifications of the B. pilosa. Our results highlighted the importance of correct species identification of materials in medicinal products.
Introduction
Bidens pilosa L. 1753 (Bartolome et al. Citation2013), classified in the Asteraceae, is a perennial and esculent plant. Parts of the plant have been widely used in traditional medicine (Chiang et al. Citation2003; Andrade-Neto et al. Citation2004; Sundararajan et al. Citation2006; Yang et al. Citation2006). At present, nearly 200 different bioactive compounds have been identified in B. pilosa, such as flavonoids, diterpenes, hydrocarbons, and terpenoids (Chang et al. Citation2004; Wang et al. Citation2010). These compounds have anti-inflammatory, anti-oxidative, immunomodulatory, and anti-ulcerogenic properties (Bartolome et al. Citation2013). The pharmacological values of the active ingredients of B. pilosa continue to attract research attention.
Bidens pilosa is native to America, but it is widely known as an introduced species to other regions of the world. Bidens pilosa has six varieties, which are separated into B. pilosa var. pilosa, var. minor, var. radiata, var. bimucronata, var. calcicola, and var. alausensis (Sherff Citation1937) based on their morphological characteristics. In China, B. pilosa as a medicinal material has the most commonly used name of ‘Mang-Chang-Cao.’ Two varieties of B. pilosa var. pilosa and var. radiata were described in FRPS (Flora Reipublicae Popularis, http://www.iplant.cn/info/Bidens%20pilosa?t=z). On the medicinal material market, the two varieties are often mixed because of similar morphology. The main difference is that the latter has glossy white flowers, but it is easy to misidentify when it is not flowering or when the glossy flowers abscise.
Alongside morphological traits, the authentication of B. pilosa can be aided by chemotaxonomy and molecular characterization (Chien et al. Citation2009). In a previous study, the DNA barcodes were used to identify three varieties, e.g. B. pilosa var. radiata, var. minor, and var. pilosa (Tsai et al. Citation2008). However, only five loci from the nuclear genome and plastome genome were selected for the variety identification. Complete plastomes with more genetic information are needed for higher species discrimination resolution.
In previous studies, two complete plastomes of B. pilosa have been deciphered (Lin et al. Citation2018; Knope et al. Citation2020). One of the plastomes was assembled from a specimen from Shaanxi, China (MN729611, labeled as Shaanxi B. pilosa), and the other was analyzed from a specimen from Hawaii, USA (MN433104, labeled as Hawaii B. pilosa). However, the plastomes from different B. pilosa specimens have not been analyzed phylogenetically. In this study, we described a novel phylogenetic relationship for all Bidens pilosa specimens and novel molecular markers for sample discrimination.
Materials
The B. pilosa specimen () were collected from the field of the Institute of Medicinal Plant Development (IMPLAD), Chinese Academy of Medical Sciences, Beijing, China (Geospatial coordinates: E116.278965, N40.041831) and identified by Professor Zhao Zhang of IMPLAD. The voucher samples of the species were deposited in the Herbarium of the institute (voucher number: IMPLADCL150801; contact person: Haimei Chen, [email protected]).
Figure 1. Panorama (A) and detail (B) photos of Bidens pilosa. The photos were shot by Liqiang Wang and the coordinates of the plant was E116.278965, N40.041831. Main identifying traits: Capitulum, margin with tongue-like flower 5–7; tongue elliptic obovate, white, 5–8 mm long; apex obtuse or notched.

Methods
Total DNA was extracted from fresh leaves using the plant genomic DNA kit (Tiangen, China) and sequenced by the Hiseq 2500 platform (Illumina, Inc., United States). The clean paired-end reads were used to assembly the complete plastome using NOVOPlasty (v. 2.7.2) (Nicolas et al. Citation2017) with default parameters. The reads were mapped to the assembled genome using Bowtie2 (v.2.0.1) (Langmead et al. Citation2009) to validate the assembly. The annotation of the plastome was performed using CPGAVAS2 (Shi et al. Citation2019).
Phylosuite (Zhang et al. Citation2020) was used to download the GenBank files of the other 45 Bidens species plastomes. Lactuca sativa and Taraxacum mongolicum from Lactuceae were designed as outgroup taxa (Supplementary Table S1). A total of 70 common CDS of the 48 plastome genome were extracted using Phylosuite and aligned using Mafft (Kazutaka and Standley Citation2013) with default parameters. The aligned CDS sequences were concatenated for reconstructing a Maximum Likelihood phylogenetic tree with the GTR + F + I + G4 model using IQ-tree2 (Quang et al. Citation2020). The bootstrap support values of the branches were calculated by using UFboot methods with 1000 replicates (Hoang et al. Citation2018).
Results
The plastome of the B. pilosa deciphered in this study (labeled as Beijing B. pilosa) is a circular DNA molecule with a total length of 150,524 bp. The reliability of genome assembly was strongly supported by the results of the mapping experiment (Figure S1). The genome has a conservative quadripartite structure, including a large-single copy (LSC) region, a small-single copy (SSC) region, and a pair of inverted repeats (IR) regions, with a length of 83,535 bp, 17,627 bp, and 24,681 bp, respectively. The GC content is 37.54%, which is lower than that of IR regions (43.06%) and higher than that of the LSC (35.58%) and SSC regions (31.23%). The plastome contains 128 genes, including 78 distinct proteins, 28 distinct tRNAs, and 4 distinct rRNA genes (). Seven protein-coding genes (rps16, rpoC1, atpF, petB, rpl2, ycf2, ndhB) contain one intron, and two genes (ycf3, clpP) contain two introns. The number of cis-splicing and trans-splicing genes were 11 (rps16, rpoC1, atpF, ycf3, clpP, petB, petD, rpl2, ndhB(×2), rpl2) and 1 (rps12) (Figure S2). Six tRNA genes (trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA and trnC-ACA) contain one intron.
Figure 2. Schematic map of overall features of the Bidens pilosa plastome. The map contains six tracks in default. From the center outward, the first track shows the dispersed repeats. The dispersed repeats consist of direct (D) and Palindromic (P) repeats, connected with red and green arcs. The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors. The colors, the type of repeat they represent, and the description of the repeat types are as follows. Black: c (complex repeat); Green: p1 (repeat unit size = 1); Yellow: p2 (repeat unit size = 2); Purple: p3 (repeat unit size = 3); Blue: p4 (repeat unit size = 4); Orange: p5 (repeat unit size = 5); Red: p6 (repeat unit size = 6). The small single-copy (SSC), inverted repeat (IRa and IRb), and large single-copy (LSC) regions are shown on the fourth track. The GC content along the genome is plotted on the fifth track. The genes are shown on the sixth track. The optional codon usage bias is displayed in the parenthesis after the gene name. Genes are color-coded by their functional classification. The transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively. The functional classification of the genes is shown in the bottom left corner.
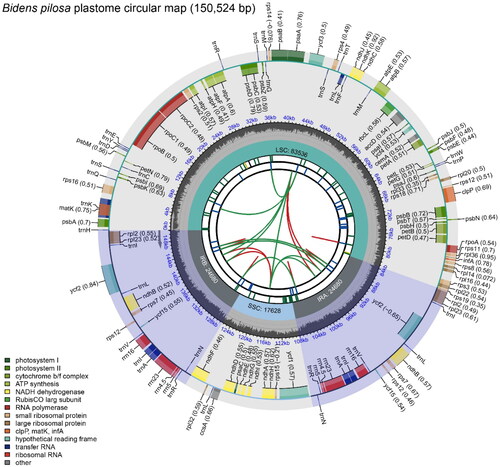
Figure 3. The Maximum-Likelihood phylogeny of Bidens pilosa and its close relatives using 70 common CDS sequences. The bootstrap values based on 1000 replicates were shown on each node in the cladogram tree. The corresponding phylogram tree was shown in the upper right corner (Dots represented Bidens species). The 45 Bidens species Heliantheae alliance were downloaded from GenBank, which were B. pilosa (MZ127828), B. pilosa (MN433104) (Knope et al. Citation2020), B. alba var. radiata (MW551952) (Wu et al. Citation2022), B. pilosa (MW551953) (Wu et al. Citation2022), B. alba var. radiata (MW551955) (Wu et al. Citation2022), B. alba var. radiata (MZ127826), B. bipinnata (MZ127827), B. alba var. radiata (MN433106), B. bipinnata (MW551954), B. bipinnata (MW551957), B. bipinnata (MW551949), B. bipinnata (MW551956), B. biternata (MW691202), B. biternata (MW551958) (Wu et al. Citation2022), B. parviflora (MW551948) (Wu et al. Citation2022), B. hillebrandiana subsp. polycephala (MN433116) (Knope et al. Citation2020), B. forbesii subsp. kahiliensis (MN433094) (Knope et al. Citation2020), B. cosmoides (MN433115) (Knope et al. Citation2020), B. forbesii subsp. forbesii (MN433028) (Knope et al. Citation2020), B. conjuncta (MN433099) (Knope et al. Citation2020), B. campylotheca subsp. pentamera (MN433107) (Knope et al. Citation2020), B. menziesii subsp. menziesii (MN433101) (Knope et al. Citation2020), B. campylotheca subsp. campylotheca (MN433029) (Knope et al. Citation2020), B. amplectens (MN433103) (Knope et al. Citation2020), B. molokaiensis (MN433097) (Knope et al. Citation2020), B. wiebkei (MN433098) (Knope et al. Citation2020), B. valida (MN433092) (Knope et al. Citation2020), B. cervicata (MN433100) (Knope et al. Citation2020), B. hawaiensis (MN433095) (Knope et al. Citation2020), B. micrantha subsp. ctenophylla (MN433114) (Knope et al. Citation2020), B. menziesii subsp. filiformis x B. micrantha subsp. ctenophylla (MN433096) (Knope et al. Citation2020), B. menziesii subsp. filiformis (MN433110) (Knope et al. Citation2020), B. micrantha subsp. kalealaha (MN433102) (Knope et al. Citation2020), B. sandvicensis subsp. confusa (MN433091) (Knope et al. Citation2020), B. sandvicensis subsp. sandvicensis (MN433093) (Knope et al. Citation2020), B. asymmetrica (MN433105) (Knope et al. Citation2020), B. macrocarpa (MN433111) (Knope et al. Citation2020), B. sandvicensis (MN433113) (Knope et al. Citation2020), B. torta (MN433112) (Knope et al. Citation2020), B. mauiensis (MN433090) (Knope et al. Citation2020), B. schimperi (MN433108) (Knope et al. Citation2020), B. pachyloma (MN433109) (Knope et al. Citation2020), B. pilosa (MN729611) (Lin et al. Citation2018), B. tripartita (MW331585) (Wu et al. Citation2022), B. pilosa (MN385242, generated in this study, labeled by bold font and a diamond), B. frondosa (MT178455) (Feifei Li et al. Citation2020). Another two species Lactuca sativa (KT272174) (Jiang et al. Citation2021) and Taraxacum mongolicum (MN164624) (Kim et al. Citation2016) from the Lactuceae served as the outgroups. Previous B. pilosa plastomes deposited in the GenBank were labeled by triangles. All B. pilosa species were located in three clades.
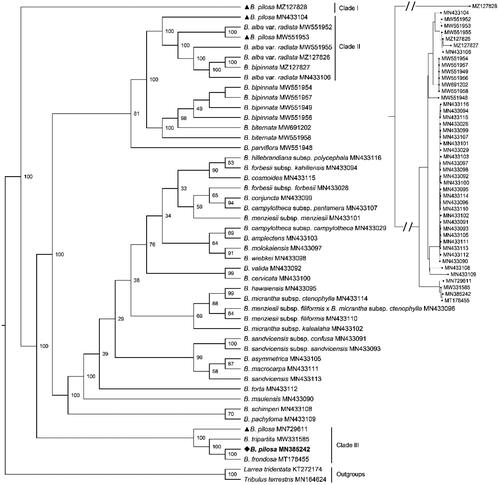
The phylogenetic analysis shown that not all B. pilosa plants formed one monophyletic clade (). They were separated into three clades. The B. pilosa (MZ127828) formed clade I with a bootstrap value of 100, which was the basic branch of the Bidens phylogenetic tree. The B. pilosa (MW551953), the Hawaii B. pilosa, and three B. alba var. radiata (MW551955, MW551952, and MN433106) formed monophyletic clade II with a bootstrap value of 100. The Beijing B. pilosa, the Shaanxi B. pilosa, the B. frondosa (MT178455), and the B. tripartita (MW331585) formed a monophyletic clade III with a bootstrap value of 100. The Beijing B. pilosa deciphered in this study was fully resolved on a branch with B. frondosa (MW331585).
Subsequently, we calculated the K2P distance between each pair of the five Bidens species in clade I, II, and III. We found the K2P distance were from 0.25 to 18.57 (Table S1). The five B. pilosa plants was also divided into three clades, which was consistent with that of the phylogenetic analysis. The K2P distance between Beijing B. pilosa and B. frondosa was 0.02. We further searched the molecular markers between the Beijing B. pilosa and the Hawaii plant. Several potential molecular markers can be found between the two plastomes using the ecoPrimer (Riaz et al. Citation2011) with core parameters of ‘-l 400 -L 500 -e 0 -3 2 -t species -T 1 -U -f -O 25’. Five pairs of conservative sequences were listed in with the highest K2P distance which could be used for designing primers for amplifying potential molecular markers.
Table 1. Conservative sequences used for designing primers for amplifying potential molecular markers between two B. pilosa plastomes.
Discussion and conclusion
In this study, we assembled and characterized a complete B. pilosa plastome using DNA next-generation data. We performed the phylogenetic analysis, calculated the K2P distance, and searched the molecular markers of different B. pilosa plants.
In the phylogenetic analysis, we revealed a large difference among the plastomes of different B. pilosa plants. Five B. pilosa plants scattered in three separated monophyletic clades, which revealed great variation of different individuals in different habitats or even possible misidentifications of the species. The K2P distance results between each pair of the five B. pilosa plants supported the results of the phylogenetic analysis. Furthermore, several potential molecular markers can be searched to distinct the Beijing and the Hawaii B. pilosa plant. The above molecular markers indicated the same results that the B. pilosa may have large variation among different individuals from different habits or even possible misidentification of the species.
The present results may indicate the B. pilosa plants from different habitats have high genetic diversity and even possible misidentifications. In the future, B. pilosa samples from various regions should be analyzed, particularly from those materials widely cultivated and used for the manufacturing of medicinal products. Reference markers based on plastomes or nuclear markers of B. pilosa will be necessary to exclude misidentifications of this species for the use of the B. pilosa materials.
Ethics statement
All samples in this study were collected with permission from the field of the Institute of Medicinal Plant Development (IMPLAD), Chinese Academy of Medical Sciences, and Peking Union Medical College, Beijing, China. This study complies with relevant institutional, national, and international guidelines and legislation.
Author contributions
The manuscript includes the contributions of all authors. Chang Liu conceived and designed this study; LiQiang Wang collected the samples; LiQiang Wang and HaiMei Chen extracted DNA for next-generation sequencing; Mei Jiang and HaiMei Chen assembled the complete plastome; Xi Wu and Qixia Yu annotated and analyzed the plastome structure; Xi Wu and Michelle Liu wrote the manuscript, calculated the K2P distance of five Bidens pilosa plants and searched novel molecular markers. Bin Wang and LiQiang Wang performed the phylogenetic analysis and revised the manuscript. All authors have read the manuscript and agreed with its contents.
Supplemental Material
Download MS Word (248.8 KB)Supplemental Material
Download MS Word (241 KB)Supplemental Material
Download MS Word (20.6 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The plastome sequence has been deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) with the accession number of MN385242 (https://www.ncbi.nlm.nih.gov/nuccore/MN385242). The associated BioProject, Bio-Sample and SRA numbers are PRJNA543381, SAMN16089020 and SRR12620715 (https://www.ncbi.nlm.nih.gov/sra/?term=SRR12620715).
Additional information
Funding
References
- Andrade-Neto VF, Brandão MGL, Oliveira FQ, Casali VWD, Njaine B, Zalis MG, Oliveira LA, Krettli AU. 2004. Antimalarial activity of Bidens pilosa L. (Asteraceae) ethanol extracts from wild plants collected in various localities or plants cultivated in humus soil. Phytother Res. 18(8):634–639.
- Bartolome AP, Villaseñor IM, Yang WC. 2013. Bidens pilosa L. (Asteraceae): botanical properties, traditional uses, phytochemistry, and pharmacology. Evid Based Complementary Altern Med. 2013(2):340215.
- Chang SL, Chang LT, Chiang YM, Hsieh RH, Tzeng CR, Wu TK, Sytwu HK, Shyur LF, Yang WC. 2004. Polyacetylenic compounds and butanol fraction from Bidens pilosa can modulate the differentiation of helper T cells and prevent autoimmune diabetes in non-obese diabetic mice. Planta Med. 70(11):1045–1051.
- Chiang LC, Chang JS, Chen CC, Ng LT, Lin CC. 2003. Anti-Herpes simplex virus activity of Bidens pilosa and Houttuynia cordata. Am J Chin Med. 31(3):355–362.
- Chien S-C, Young PH, Hsu Y-J, Chen C-H, Tien Y-J, Shiu S-Y, Li T-H, Yang C-W, Marimuthu P, Tsai LF-L, et al. 2009. Anti-diabetic properties of three common Bidens pilosa variants in Taiwan. Phytochemistry. 70(10):1246–1254.
- Feifei Li JZ, Xiaoyan L, Kexiao G, Caiyun Z. 2020. The complete chloroplast genome of an invasive herb Bidens frondosa L. (Asteraceae). Mitochondrial DNA B Res. 5(2):1769–1770.
- Hoang DT, Chernomor O, Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522.
- Jiang M, Li Y, Chen H, Wang B, Liu C. 2021. Comparative and phylogenetic analysis of the complete chloroplast genome sequences of Lactuca raddeana and Lactuca sativa. Mitochondrial DNA B Res. 6(4):1498–1506.
- Kazutaka K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kim JK, Park JY, Lee YS, Woo SM, Park HS, Lee SC, Kang JH, Lee TJ, Sung SH, Yang TJ. 2016. The complete chloroplast genomes of two Taraxacum species, T. platycarpum Dahlst. and T. mongolicum Hand.-Mazz. (Asteraceae). Mitochondrial DNA B Res. 1(1):412–413.
- Knope ML, Renee BM, Datlof EM, Gallaher TJ, Johnson MA. 2020. Insights into the evolutionary history of the Hawaiian bidens (Asteraceae) adaptive radiation revealed through phylogenomics. J Hered. 111(1):1–19.
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10(3):R25.
- Lin Y-X, Duan R-Y ,Tan Z-X, Ma Y-H, Wu H. 2018. The complete chloroplast genome of the invasive and Cd-hyperaccumulator herb Bidens pilosa L. (Asteraceae). Mitochondrial DNA Part B. 3(2):746–747.
- Nicolas D, Patrick M, Guillaume S. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Quang MB, Schmidt HA, Olga C, Dominik S, Woodhams MD, Arndt VH, Robert L. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Riaz T, Shehzad W, Fau-Viari A, Viari A, Pompanon F, Taberlet P, Coissac E. 2011. ecoPrimers: inference of new DNA barcode markers from whole genome sequence analysis. Nucleic Acids Res. 39(21):e145.
- Sherff EE. 1937. The genus Bidens. Int J Plant Sci. 11:412–461.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Sundararajan P, Dey A, Smith A, Doss AG, Natarajan S. 2006. Studies of anticancer and antipyretic activity of Bidens pilosa whole plant. African Health Sciences. 6(1):27–30.
- Tsai LC, Wang JC, Hsieh HM, Liu KL, Linacre A, Lee CI. 2008. Bidens identification using the noncoding regions of chloroplast genome and nuclear ribosomal DNA. Forensic Sci Int Genet. 2(1):35–40.
- Wang NL, Wang J, Yao XS, Kitanaka S. 2010. Two neolignan glucosides and antihistamine release activities from Bidens parviflora Willd. Chem Pharm Bull. 38(3):1190–1192.
- Wu L, Nie L, Guo S, Wang Q, Wu Z, Lin Y, Wang Y, Li B, Gao T, Yao H. 2022. Identification of medicinal Bidens plants for quality control based on organelle genomes. Front Pharmacol. 13:842131.
- Yang H-L, Chen S-C, Chang N-W, Chang J-M, Lee M-L, Tsai P-C, Fu H-H, Kao W-W, Chiang H-C, Wang H-H, et al. 2006. Protection from oxidative damage using Bidens pilosa extracts in normal human erythrocytes. Food Chem Toxicol. 44(9):1513–1521.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.
