Abstract
Mystus gulio Hamilton, also called long whiskers catfish, is an endemic fish and a common food in some Asian countries. In this study, the complete mitochondrial genome of M. gulio was sequenced using the MinION system (Oxford Nanopore Technologies). The mitochondrial genome is 16,518 bp in length (41.1% [G + C] content) and consists of 13 protein-coding genes, 22 transfer RNA genes, and two ribosomal RNA genes. The results of phylogenetic analysis inferred from whole mitochondrial genomes of Mystus and related species in the family Bagridae revealed that M. gulio is closely related to Mystus cavasius.
Introduction
Mystus gulio Hamilton 1822, which belongs to the genus Mystus of the family Bagridae, order Siluriformes, is a small fish distributed in coastal waters from India to Indonesia and Vietnam (Talwar and Jhingran, Citation1991). It was assessed as a vulnerable species, and its population size is decreasing (Hossain et al. Citation2015). This fish, also known as “cá chốt” in Vietnamese, “nuna tengra” in Bengali, and “pla kayeng noo” in Thai (Froese and Pauly, Citation2022), has long been used in traditional food and has high commercial value in coastal fisheries of India, Bangladesh, Thailand, and Vietnam (Rahman et al. Citation2020). M. gulio is classified as a species of least concern in the International Union for Conservation of Nature Red List of Threatened Species; however, it is facing destructive fishing pressure, decreasing populations, habitat loss, and threats due to climate change (Hossain et al. Citation2015; Ng et al. Citation2019; Rahman et al. Citation2020). Although studies of the morphology, phylogeny, and reproductive biology of M. gulio have been conducted, there have been no extensive genomic studies (i.e. complete mitochondrial genome/mitogenome) of M. gulio (Gupta Citation2014; Kumar et al. Citation2019; Liu et al. Citation2019; Widayanti et al. Citation2021; Hashimoto et al. Citation2022). Therefore, we characterized the complete mitochondrial genome of M. gulio and analyzed its phylogenetic relationship within the family Bagridae. The new genomic data of M. gulio will provide fundamental information for further studies on the population genetics, evolution, and molecular markers of this species.
Materials and methods
Samples (whole dead organisms) of M. gulio were collected in the Mekong Delta region, Can Tho City, Vietnam (10° 0′ 22.691″ N, 105° 45′ 3.311″ E). No specific permission was required to collect this species in Vietnam. The samples were stored at −80 °C and deposited at NTT Hi-Tech Institute – Nguyen Tat Thanh University (determined by Dr. Do Hoang Dang Khoa, email: [email protected]) under voucher number NTTU-202210-A001 (). Total genomic DNA was extracted from liver tissue by the salt-out method (El-Ashram et al. Citation2016). To isolate the mitochondrial genome sequence, a long-range PCR approach was conducted using LongAmp® Taq DNA Polymerase (#M0323S; New England Biolabs). The complete mitochondrial genome sequence of Mystus cavasius Hamilton 1822 (GenBank accession number NC_030187), Mystus rhegma Fowler 1935 (GenBank accession number NC_023223), and Mystus vittatus Bloch 1794 (GenBank accession number NC_032082) were used to design two primer pairs using Primer3 (Untergasser et al. Citation2012): 16SF (5′-CGCCTGTTTATCAAAAACAT-3′)/CytobR (5′-GGAATGCGAAGAATCGTGTT-3′) and trnLF (5′-CTCTTGGTGCAANTCCAAG-3′)/16SR (5′-CCGGTCTGAACTCAGATCACGT-3′), which amplified PCR products of 13 kb and 7 kb, respectively. The PCR protocol consisted of 2 min at 94 °C, followed by 35 cycles of 10 s at 94 °C, 30 s at 50 °C, and 6 min at 65 °C, with an extension step of 2 min at 65 °C. The PCR products were checked by 1% agarose gel electrophoresis. The amplicons were pooled before purification using the Monarch PCR & DNA cleanup kit (#T1030, New England Biolabs). The purified DNA was then used to prepare a sequencing library using Native Barcoding Expansion 1-12 (#EXP-NBD104, Oxford Nanopore Technologies) and the Ligation Sequencing Kit (SQK-LSK109, Oxford Nanopore Technologies) according to the manufacturer’s instructions. The prepared library was loaded into the MinION device with R9.4.1 flow cells (#FLO-MIN106, Oxford Nanopore Technologies) for sequencing. MinKNOW was used to monitor the sequencing process. Raw data (in fast5 format) were converted to fastq format using Guppybasecaller v5.0.7 (Wick et al. Citation2019). The fastq data were then assembled and the mitochondrial genome sequence of M. gulio was analyzed using Geneious Prime v2022.1 (https://www.geneious.com) with the mitochondrial genome of M. cavasius as a reference. The complete mitochondrial genome (average coverage depth = 257x) of M. gulio was annotated using GeSeq (Tillich et al. Citation2017). The complete mitochondrial sequence was deposited in GenBank under accession number OP650114. For phylogenetic analysis, the complete mitochondrial genomes of 45 species belonging to the Bagridae family and one species of Gymnotidae (Gymnotus sylvius Albert and Fernandes-Matioli 1999) was included as an outgroup. The sequences were aligned using MUSCLE v5 (Edgar Citation2004). The aligned data were tested for the best substitution model using jModelTest 2 (Darriba et al. Citation2012). The results showed that GTR + I + G is the best model for the data matrix of Mystus and related species. The maximum likelihood (ML) method was used for phylogenetic analysis with 1000 bootstrap replicates using IQ-TREE2 (Minh et al. Citation2020). For Bayesian inference (BI) analysis, the data matrix was analyzed using two Markov chain Monte Carlo runs for one million generations in MrBayes v3.2.7 (Huelsenbeck and Ronquist Citation2001). The tree was sampled every 1000 generations and 25% were discarded as burn-in. BI analysis was performed when the split frequency was lower than 0.01. The phylogenetic tree was modified and illustrated using Figtree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/).
Results
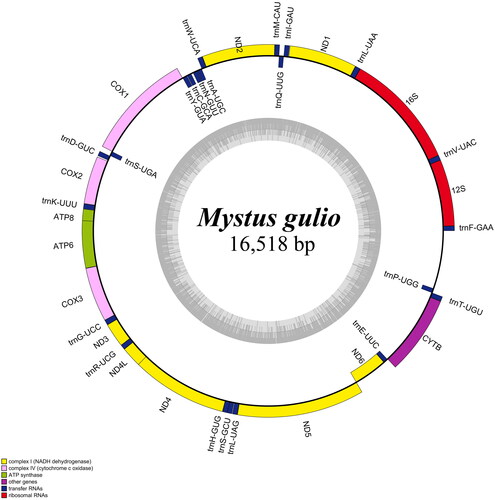
The mitogenome of M. gulio was 16,518 bp in length (GenBank accession number OP650114) and included 22 transfer RNA (tRNAs) genes (tRNAAla, tRNAArg, tRNAAsn, tRNAAsp, tRNACys, tRNAGln, tRNAGlu, tRNAGly, tRNAHis, tRNAIle, tRNALeu (2×), tRNALys, tRNAMet, tRNASer (2×), tRNAPhe, tRNAPro, tRNAThr, tRNATrp, tRNATyr, and tRNAVal), 13 protein-coding genes (ND1, ND2, COX1, COX2, ATP8, ATP6, COX3, ND3, ND4L, ND4, ND5, ND6, and CYTB), and two rRNAs (12S rRNA and 16S rRNA; ). Among the protein-coding genes, ND2, COX2, COX3, ND3, ND4, and CYTB might have the fulfillment of incomplete stop codon T by posttranscriptional polyadenylation with poly-A tail (Ojala et al. Citation1981). The percentages of A, C, T, and G in the mitogenome were 31.9% (5265 nucleotides), 26.1% (4308 nucleotides), 27.0% (4459 nucleotides), and 15.1% (2486 nucleotides), respectively, with an [A + T] content of 58.9%. The gene content and order of the new mitogenome were similar to those of the published mitogenomes of other Mystus species. Specifically, the pairwise identities of mitogenomes of M. gulio compared with M. cavasius, M. Rhegma, and M. vittatus were 89.2%, 87.5%, and 81.6%, respectively.
Figure 2. Map of the mitochondrial genome of M. gulio. The genes transcribed counterclockwise are located outside the circle, while genes transcribed clockwise are inside the circle. The dark gray area in the inner circle indicates the GC content of the mitochondrial genome, whereas the AT content is represented by a light gray area. Genes were classified into different functional groups that were shown in different color blocks.
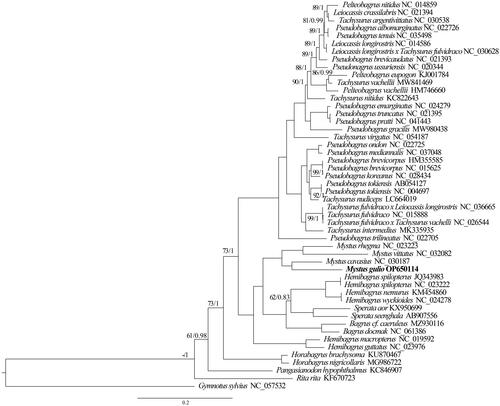
Phylogenetic analysis inferred from 46 complete mitogenomes revealed the same topology between the ML and BI methods (). The monophyly of Mystus species was supported by high support values (bootstrap (BS)=100/posterior probability (PP)=1). In contrast, the paraphyly and polyphyly of Hemibagrus, Pseudobagrus, Leiocassis, and Tachysurus were found. Both BI and ML analyses supported a close relationship between M. gulio and M. cavasius.
Discussion and conclusion
In this study, the complete mitochondrial genome of M. gulio was sequenced and characterized. The newly reported mitochondrial genome is similar in gene content and order to those of other Mystus species (Duan et al. Citation2019). In addition, the new genomic data revealed phylogenetic relationships between M. gulio and related species in Bagridae. Specifically, M. gulio is closely related to M. Casavius, which was reported in previous studies. The newly reported mitochondrial genome represents essential information for further studies on the evolutionary history, phylogeography, and conservation of M. gulio.
Author contributions
Hoang Dang Khoa Do conceive the conception and design; Nguyen Hoang Danh and Hoang Dang Khoa Do collected and determined the samples; Nguyen Hoang Danh, Vu Minh Thiet, and Hoang Dang Khoa Do conducted the experiments, analyzed the data, and wrote the draft manuscript; Vu Minh Thiet and Hoang Dang Khoa Do revised the draft manuscript. All authors agreed to the final form of this manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data generated in this study are openly available in the GenBank database https://www.ncbi.nlm.nih.gov/genbank/under the accession number OP650114. The associated BioProject, SRA, and BioSample numbers are: PRJNA889671, SRR21869731, and SAMN31249233 respectively
Additional information
Funding
References
- Darriba D, Taboada G, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.
- Duan S, Pan H, Peng Z. 2019. The complete mitochondrial genome of Mystus rhegma (Teleostei: siluriformes) and its phylogenetic position. Mitochondrial DNA Part B. 4(1):1788–1789.
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinf. 5(1):113.
- Froese R, Pauly D. 2022. FishBase. World Wide Web electronic publication. Facilitated by FishBase Consortium (Accessed date 2022 Agust 08). www.fishbase.org.
- Gupta S. 2014. Morphology, growth pattern, feeding and reproductive biology of Mystus gulio (Hamilton-Buchanan, 1822) (Siluriformes: bagridae). Int J Aquat Biol. 2(4):201–205.
- Hashimoto S, Kakehashi R, Mori T, Kambayashi C, Kanao S, Kurabayashi A. 2022. The complete mitochondrial genome of a Bagrid Catfish, Tachysurus nudiceps, and its phylogenetic implications for the classification of the Bagrid Genera. Mitochondrial DNA Part B Resour. 7(4):606–608.
- El-Ashram S, Al Nasr I, Suo X. 2016. Nucleic acid protocols: extraction and optimization. Biotechnol Rep . 12:33–39.
- Hossain MY, Islam R, Hossen MA, Rahman O, Hossain MA, Islam MA, Alam MJ. 2015. Threatened fishes of the world: Mystus gulio (Hamilton, 1822) (Siluriformes: Bagridae). CJF. 73(1):43–45.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755.
- Liu Y, Wu P-D, Zhang D-Z, Zhang H-B, Tang B-P, Liu Q-N, Dai L-S. 2019. Mitochondrial genome of the Yellow Catfish Pelteobagrus fulvidraco and insights into Bagridae phylogenetics. Genomics. 111(6):1258–1265.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Ng HH, de Alwis Goonatilake S, Fernado M, Kotagama O. 2019. Mystus gulio. The IUCN Red List of Threatened Species. 2019: e.T144201289A144201789. Facilitated by International Union for Conservation of Nature and Natural Resources (Accessed 2022 July 17). https://dx.doi.org/10.2305/IUCN.UK.2019-3.RLTS.T144201289A144201789.en
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290(5806):470–474.
- Kumar P, Biswas G, Ghoshal TK, Kailasam M, Christina L, Vijayan KK. 2019. Current knowledge on the biology, captive breeding and aquaculture of the brackishwater catfish, Mystus gulio (Hamilton, 1822): a review. Aquaculture. 499:243–250.
- Rahman O, Hossain MY, Islam MA, Rahman MA, Khatun D, Parvin Most F, Sarmin Most S, Tanjin S, Rahman MA, Mawa Z, et al. 2020. Life-history traits of long Whisker Catfish Mystus Gulio (Siluriformes: bagridae) in the coastal water (Maloncho River) of Southern Bangladesh. Pak J Mar Sci. 29(2):99–114.
- Talwar PK, Jhingran AG. 1991. Inland Fishes of India and Adjacent Countries. New Delhi: Oxford-IBH Publishing Co. Pvt. Ltd. p560.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. 2012. Primer3–new capabilities and interfaces. Nucleic Acids Res. 40(15):e115.
- Wick RR, Judd LM, Holt KE. 2019. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol. 20(1):129.
- Widayanti R, Kusumaastuti KA, Novi JM, Adani FK, Gultom CRP, Prastiti AD, Nugroho HA, Pakpahan S. 2021. Genetic variation and phylogenetic analysis of Indonesian indigenous catfish (baung fish) based on mitochondrial 12S rRNA gene. Vet World. 14(3):751–757.