Abstract
Amsonia elliptica (Apocynaceae), endangered species in Korea, is a perennial herb that is economically important as traditional medicine and used as ornamentals. Natural populations of this species are facing extinction due to small population size and isolated distribution. Here, we report the complete chloroplast (cp) genome of A. elliptica using Illumina HiSeq sequencing and its phylogenetic position in subfamily Rauvolfioideae based on 20 Apocynaceae cp genomes. The cp genome of A. elliptica was 154,242 bp in length with a pair of inverted repeats of 25,711 bp, separated by large single-copy and small single-copy regions of 85,382 bp and 17,438 bp, respectively. Our phylogenomic analyses revealed that A. elliptica was closely related to Rhazya stricta in Rauvolfioideae (Apocynaceae).
Introduction
Amsonia elliptica (Thunb.) Roem. & Schult. 1819 (Apocynaceae) is a perennial herb distributed in China, Japan, and Korea and economically important as ornamentals and medicine (Li et al. Citation1995; Lee Citation2018). In Korea, this species occurs in the grass area along the coast of the islands and it is now only observable at four localities. Therefore, A. elliptica is considered to be endangered due to its isolated distribution and small population size (Korea National Arboretum Citation2021). Information of chloroplast (cp) genomes has been extensively applied to understand the phylogenetic relationship and conservation genetics of an endangered species (Jung et al. Citation2021; Wei and Li Citation2022), while studies regarding this species is lacking. Here, we provide the complete cp genome of A. elliptica using next generation sequencing technology.
Materials and methods
Plant materials and DNA extraction
Total genomic DNA was extracted from silica-dried leaf material of a single individual of A. elliptica from the Yaksan Island, Wando-gun, Jeollanam-do, South Korea (34.3772N, 126.9314E; ) using a DNeasy Plant kit (Qiagen Co., Valencia, CA). The voucher specimen was deposited at the herbarium of Honam National Institute of Biological Resources (HIBR (https://hnibr.re.kr/); contact person Yongsung Kim, [email protected]; ) under the voucher no. HNIBRVP14274.
Figure 1. The habit and voucher specimen of Amsonia elliptica. (a) Habit of A. elliptica (photo by Bo-Mi Nam, 30 April 2022. Yaksan Island, Wando-gun, South Korea). Amsonia elliptica is a perennial herb and up to 80 cm tall. Leaf blade elliptic, base and apex acuminate; corolla bluish, salverform. (b) The voucher specimen of A. elliptica with a pair of follicles (collected by C. Kim, 4 September 2022, Yaksan Island, Wando-gun, South Korea; herbarium number: HNIBRVP14274 [HIBR]).
![Figure 1. The habit and voucher specimen of Amsonia elliptica. (a) Habit of A. elliptica (photo by Bo-Mi Nam, 30 April 2022. Yaksan Island, Wando-gun, South Korea). Amsonia elliptica is a perennial herb and up to 80 cm tall. Leaf blade elliptic, base and apex acuminate; corolla bluish, salverform. (b) The voucher specimen of A. elliptica with a pair of follicles (collected by C. Kim, 4 September 2022, Yaksan Island, Wando-gun, South Korea; herbarium number: HNIBRVP14274 [HIBR]).](/cms/asset/d362a627-04f4-461a-9bb9-b65cbe626156/tmdn_a_2192834_f0001_c.jpg)
Genome sequencing, assembly, and annotation
Genomic DNA was sequenced using an Illumina HiSeq sequencer (Illumina, San Diego, CA). The complete cp genome was assembled via NOVOPlasty 4.3 (Dierckxsens et al. Citation2017). The depth of the reads mapping was verified and visualized by Geneious Prime 2022.0.1 (Biomatters Ltd., Auckland, New Zealand). The genome was automatically annotated using CPGAVAS2 (Shi et al. Citation2019) and then adjusted and confirmed with Geneious Prime 2022.0.1. The tRNA sequences were also confirmed by tRNAscan-SE (Lowe and Eddy Citation1997). Schematic representation of cp genome structure and detailed structures of the cis-splicing and trans-splicing genes were illustrated using the CPGView (http://www.1kmpg.cn/cpgview; Liu et al. Citation2023). The complete cp genome sequence of A. elliptica was submitted to GenBank (accession no. OP474151).
Phylogenetic analysis
To determine the phylogenetic relationship, in addition to A. elliptica, we included 17 species (19 accessions) from the subfamily Rauvolfioideae of Apocynaceae. Within the subfamily, two species (Aspidosperma cruentum Woodson and Strempeliopsis strempelioides (Griseb.) Benth. ex B.D.Jacks.) from Aspidospermateae were designated as outgroups based on previous phylogenetic studies (Simões et al. Citation2007; Nazar et al. Citation2019). Multiple-sequence alignment of the 76 cpDNA coding regions was performed using MAFFT with the default alignment parameters and then edited manually (Katoh et al. Citation2019). The phylogenomic reconstruction was performed with maximum-likelihood (ML) and Bayesian inference (BI) methods using the concatenated 76 cpDNA coding regions. We used the IQ-TREE web server (http://iqtree.cibiv.univie.ac.at/) to make the ML tree with TVM + F+R2. The BI phylogram was reconstructed using MrBayes v.3.2.6 (Ronquist et al. Citation2012) with the following parameters: nst = 6, rates = invgamma, ngen = 1,000,000, samplefreq = 1000, burn-in = 25%. Bootstrap support (BS; 1000 replicates) and posterior probability (PP) were calculated to estimate robustness for each clade.
Results
General feature of the chloroplast genome of A. elliptica
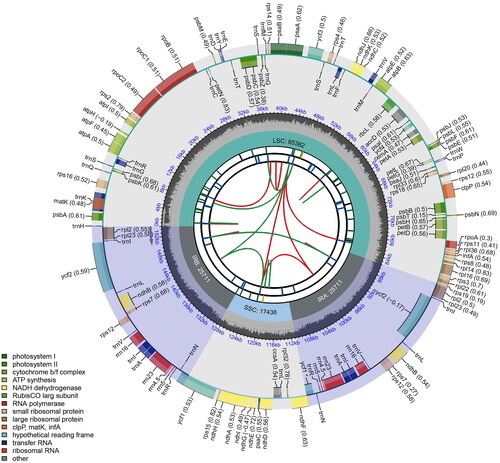
The average and minimum read mapping depth of assembled genome of A. elliptica were 1640.9 X and 421 X, respectively (Figure S1). The cp genome of A. elliptica consisted 154,242 bp with the typical conserved ‘quadripartite’ structure, with a pair of inverted repeats (IR; 25,711 bp) separated by large single-copy (LSC; 85,382 bp) and small single-copy (SSC; 17,438 bp) regions (). The cp genome of A. elliptica included 85 protein-coding genes, 37 tRNA genes, and eight rRNA genes. One pseudogene (ycf1) is located at the IR-SSC junction. Totally, 11 cis-splicing and one trans-splicing genes were detected (Figure S2). The overall GC content is 37.2%, and in the LSC, SSC, and IR regions are 35.9%, 32.4%, and 43.3%, respectively.
Figure 2. Schematic map of overall features of complete chloroplast genome of Amsonia elliptica. The map contains six circles. From the center outward, the first circle shows the distributed repeats connected with red (forward direction) and green (reverse direction) arcs. The second circle shows tandem repeats marked with short bars. The third circle shows the short tandem repeats of microsatellite sequences as short bars with different colors. The fourth circle displays the sizes of large single-copy (LSC), small single-copy (SSC), and inverted repeat (IRa and IRb) regions. The fifth circle illustrates the distribution of GC contents along the chloroplast genome (dark grey, GC content; light grey, background). The sixth circle displays the genes with colored boxes.
Phylogenetic relationship of A. elliptica
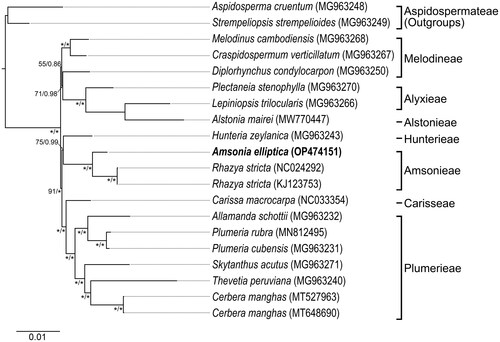
The ML and BI trees, based on 76 cpDNA genes from Amsonia and related taxa, yield identical topologies (data not shown). Within Rauvolfioideae, A. elliptica was sister to Rhazya stricta Decne. with strong support (BP = 100%; PP = 1.00) forming the robust monophyly of the tribe Amsonieae ().
Figure 3. Maximum-likelihood phylogram inferred from 76 cpDNA coding regions of 20 cp genomes from Rauvolfioideae (Apocynaceae). Numbers near nodes indicate support values (maximum likelihood (BS)/Bayesian posterior probability (PP)); an asterisk indicates that a node BS = 100% for maximum-likelihood analysis and PP = 1.00 for Bayesian’s inference analysis. The scale bar shows the number of substitutions per site. Tribes of Rauvolfioideae classification follow Endress et al. (Citation2014). The target species was marked in bold. The following sequences were used: Aspidosperma cruentum MG963248 (Fishbein et al. Citation2018), Strempeliopsis strempelioides MG963249 (Fishbein et al. Citation2018), Melodinus cambodiensis MG963268 (Fishbein et al. Citation2018), Craspidospermum verticillatum MG963267 (Fishbein et al. Citation2018), Diplorhynchus condylocarpon MG963250 (Fishbein et al. Citation2018), Plectaneia stenophylla MG963270 (Fishbein et al. Citation2018), Lepiniopsis trilocularis MG963266 (Fishbein et al. Citation2018), Alstonia mairei MW770447 (unpublished), Hunteria zeylanica MG963243 (Fishbein et al. Citation2018), Amsonia elliptica OP474151 (this study), Rhazya stricta NC024292 (Park et al. Citation2014), KJ123753 (unpublished), Carissa macrocarpa NC033354 (Jo et al. Citation2017), Allamanda schottii MG963232 (Fishbein et al. Citation2018), Plumeria rubra MN812495 (Wang et al. Citation2020), Plumeria cubensis MG963231 (Fishbein et al. Citation2018), Skytanthus acutus MG963271 (Fishbein et al. Citation2018), Thevetia peruviana MG963240 (Fishbein et al. Citation2018), and Cerbera manghas MT527963 (Liao et al. Citation2020), MT648690 (unpublished).
Discussion and conclusions
In this study, the cp genome of A. elliptica was first sequenced, assembled, and annotated. The cp genomes are highly conserved in angiosperms with the respect to the structure and gene content (Daniell et al. Citation2016). This conservative tendency was observed in newly sequenced Amsonia cp genome, compared to Rhazya of Amsonieae. Both the cp genomes of Amsonia and Rhazya showed typical quadripartite structures and included 85 protein-coding genes, 37 tRNA genes, and eight rRNA genes (Park et al. Citation2014).
Based on the phylogenetic analysis of Rauvolfioideae, the sister group of Amsonia is Rhazya with strong support (). This result is in line with a previous study of the molecular phylogenetic relationships of Apocynaceae (Nazar et al. Citation2019). The monophyly of Amsonia-Rhazya is also supported by having similar pollen grains including exine ultrastructure (Nilsson Citation1990). Despite the molecular and palynological distinctiveness of Amsonieae as a tribe of Rauvolfioideae, its sister group is unclear based on our current sampling that was not representative of the entire tribes. Thus, the sister of Amsonieae needs to be evaluated by a broad taxon sampling scheme of Rauvolfioideae.
Our complete cp genome data of A. elliptica may be useful in assessing the genetic diversity, population structure, and phylogeographic history of this species, and will help to infer its molecular identification, thereby providing a guideline for conservation.
Ethical approval
Field works have complied with local legislation and appropriate permissions/license were granted while taking samples from a protected land. The process and purpose of this experimental research were in line with the rules and regulations of our institute. There are no ethical issues or other conflicts of interest in this study.
Author contributions
CK and TD conceived and designed the research. B-MN and CK collected the plant materials. YK and IK performed the experiments and analyzed the data. YK, IK, and B-MN wrote the draft. CK and TD revised the draft. All authors approved the final version of the manuscript and agreed to be accountable for all aspects of the work.
Supplemental Material
Download JPEG Image (1.1 MB)Supplemental Material
Download JPEG Image (511.6 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome data that supported the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. OP474151. The associated BioProject, SRA, and Bio-sample numbers are PRJNA885521, SRR21763746, and SAMN31098060, respectively.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Daniell H, Lin C-S, Yu M, Chang W-J. 2016. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17(1):134.
- Endress ME, Liede-Schumann S, Meve U. 2014. An updated classification for Apocynaceae. Phytotaxa. 159(3):175–194.
- Fishbein M, Livshultz T, Straub SCK, Simões AO, Boutte J, McDonnell A, Foote A. 2018. Evolution on the backbone: Apocynaceae phylogenomics and new perspectives on growth forms, flowers, and fruits. Am J Bot. 105(3):495–513.
- Jo S, Kim H-W, Kim Y-K, Cheon S-H, Kim K-J. 2017. The complete plastome sequence of Carissa macrocarpa (Eckl.) A.DC. (Apocynaceae). Mitochondrial DNA B Resour. 2(1):26–28.
- Jung J, Kim C, Kim J-H. 2021. Insights into phylogenetic relationships and genome evolution of subfamily Commelinoideae (Commelinaceae Mirb.) inferred from complete chloroplast genomes. BMC Genomics. 22(1):231.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Korea National Arboretum. 2021. National red list of vascular plant in Korea. Pocheon (Korea): Korea National Arboretum Press.
- Lee ST. 2018. Amsonia Walter. In: Flora of Korea Committee, editors. The genera of vascular plants of Korea. Seoul (Korea): Hong-Reung Science Publishing Co.; p. 1027–1028.
- Li P-T, Leeuwenberg AJM, Middleton DJ. 1995. Amsonia Walter. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 16. Beijing (China): Science Press; p. 156.
- Liao M, Wei X-F, Ding H-P, Tang G-D. 2020. The complete chloroplast genome of the highly poisonous plant Cerbera manghas L. (Apocynaceae). Mitochondrial DNA B Resour. 5(3):3084–3085.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.
- Nazar N, Clarkson JJ, Goyder D, Kaky E, Mahmood T, Chase MW. 2019. Phylogenetic relationships in Apocynaceae based on nuclear PHYA and plastid trnL-F sequences, with a focus on tribal relationships. Caryologia. 72(1):55–81.
- Nilsson S. 1990. Taxonomic and evolutionary significance of pollen morphology in the Apocynaceae. In: Hesse M, Ehrendorfer F, editors. Morphology, development, and systematic relevance of pollen and spores. Plant systematics and evolution. Vol. 5. Vienna: Springer; p. 91–102.
- Park S, Ruhlman TA, Sabir JSM, Mutwakil MHZ, Baeshen MN, Sabir MJ, Baeshen NA, Jansen RK. 2014. Complete sequences of organelle genomes from the medicinal plant Rhazya stricta (Apocynaceae) and contrasting patterns of mitochondrial genome evolution across asterids. BMC Genomics. 15(1):405.
- Ronquist F, Teslenko M, Van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Simões AO, Livshultz T, Conti E, Endress ME. 2007. Phylogeny and systematics of the Rauvolfioideae (Apocynaceae) based on molecular and morphological evidence. Ann MO Bot Gard. 94(2):268–297.
- Wang D-L, Liu Y-Y, Tian D, Yu L-Y, Gui L-J. 2020. Characterization of the complete chloroplast genome of Plumeria rubra cv. Acutifolia (Apocynaceae). Mitochondrial DNA B Resour. 5(1):927–928.
- Wei R, Li Q. 2022. The complete chloroplast genome of endangered species Stemona parviflora: insight into the phylogenetic relationship and conservation implications. Genes. 13(8):1361.
