Abstract
The mitochondrial genome of the tachinid fly Peleteria iavana (Wiedemann, 1819), which belongs to the family Tachinidae, was obtained for the first time using high-throughput sequencing techniques. The complete mitochondrial genome is 15,697 bp in size and consists of 13 protein-coding genes (PCGs), two ribosomal RNA genes, 22 transfer RNAs and a non-coding control region. The nucleotide composition biases A and T, the overall A + T percentage is up to 78.9% of the entire mitogenome. A phylogenetic analysis of 30 species within the family Tachinidae suggested that P. iavana is most closely related to (Janthinomyia sp.+Lydina aenea). The P. iavana mitochondrial genome will be a fundamental resource for understanding the molecular phylogenetic relationships of the species-rich subfamily Tachininae of Tachinidae.
Introduction
The Tachininae is the second most speciose subfamily in Tachinidae, with 2,746 described species worldwide (O’Hara et al. Citation2020). As a parasitoid, it is an important natural enemy in most terrestrial ecological communities (Stireman et al. Citation2006). Genus Peleteria is a representative member of Tachininae, including 4 subgenera and 124 species in the world, while 2 species occur in Guizhou Province, China (O’Hara et al. Citation2009; Citation2020). Peleteria iavana (Wiedemann, 1819) spreads throughout the Palearctic, Oriental, Afrotropical and Australasian Regions (O’Hara and Henderson Citation2020), and it is considered a native species of China (O’Hara et al. Citation2009). In this study, the mitogenome data of P. iavana is sequenced and annotated as the first mitogenome of genus Peleteria, and the phylogenetic placement of P. iavana in the family Tachinidae is explored.
Materials and methods
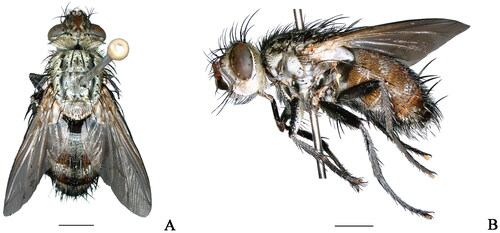
The specimen () was collected by sweeping collection from Beipanjiang Town, Zhenfeng County, Guizhou Province, China (25°31′48″N, 105°45′36″E) on 26 July 2021 and was deposited at the Key Laboratory of Medical Insects of Guizhou Medical University (https://www.gmc.edu.cn/, Jiayu Liu, [email protected]) under the voucher number PI20210726. Photographs were taken with the KEYENCE VHX-6000 system. The sequencing library was established and then used the Illumina Hiseq PE150 platform for whole genome sequencing. The raw reads were assembled using Mitoz 2.4-alpha with default parameters (Meng et al. Citation2019). The assembled contigs were aligned with the previously published mitochondrial genomes of Hamaxiella brunnescens (MW256712.1) and Janthinomyia sp. (MK644822.1) using Geneious Prime 2020.2.2 (Kearse et al. Citation2012). Annotation of the mitogenome, including gene prediction and non-coding RNA analysis, was conducted using MITOS2 Web Server (http://mitos2.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013) and tRNAscan-SE version 1.21 (Schattner et al. Citation2005). Subsequently, the mitogenome data were submitted to GenBank database through NCBI.
Figure 1. Adult of Peleteria iavana (Wiedemann, 1819). (A) Dorsal view; (B) Lateral view. Scale bars: 2.0 mm. (photography by Peng Zhang).
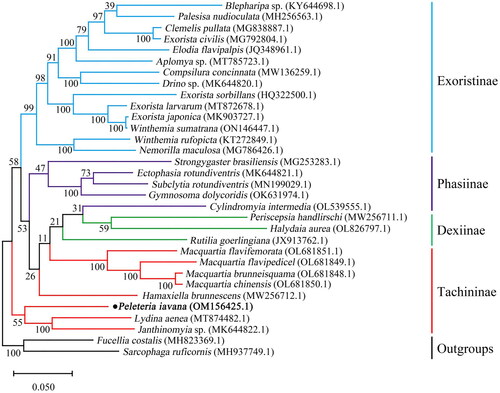
To reveal the phylogenetic position of P. iavana, the dataset of 13 PCGs of previously reported 29 tachinid species were downloaded and their GenBank accession numbers were provided in (Nelson et al. Citation2012; Shao et al. Citation2012; Zhao et al. Citation2013; Li et al. Citation2017; Hou et al. Citation2018, Citation2019; Pei et al. Citation2019; Seo et al. Citation2019; Luo et al. Citation2021; Yan et al. Citation2021; Li et al. Citation2022; Wang, Zhi, Zhang et al. Citation2022; Wang, Zhi, Yao et al. Citation2022). Additionally, Fucellia costalis from Anthomyiidae and Sarcophaga ruficornis from Sarcophagidae were chosen as outgroups. Preliminary multiple sequence alignments were performed using the Clustal W in MEGAX 10.2.6 (Kumar et al. Citation2018). The phylogenetic tree was reconstructed in the maximum-likelihood (ML) method with 1,000 bootstrap repeats using MEGA X 10.2.6 (Kumar et al. Citation2018).
Results
The 4.43 Gb raw data was yielded by the high-throughput sequencing techniques with 12,926,828 reads, and the 4.29 Gb (96.84%) clean data was obtained after filtering. The complete mitochondrial genome of P. iavana was assembled using 594,117 reads, and the average depth of coverage was assessed at 4561.6 X (Supplementary Figure 1). The average coverage depth for two regions nucleotides 14,853–14,977 (A + T ratio: 94.4%) and nucleotides 15,147–15,390 (A + T ratio: 93.5%) was 503.1 X (max: 635 X, min: 426 X) and 306.8 X (max: 632 X, min: 138 X), respectively. The reason that the two regions had relatively low coverage depth in the mitochondrial genome may be that the control region had a higher AT ratio and more repeat sequences. The complete circular mitogenome of P. iavana (GenBank accession number: OM156425.1) was 15,697 bp in length, with the base composition as follows: A (40.5%), T (38.4%), G (9.0%), and C (12.1%). The nucleotide composition was AT-biased (A + T ratio: 78.9%). It contained 13 protein-coding genes (PCGs), two rRNA genes, 22 tRNA genes and one non-coding control region, which was a typical configuration of the tachinid mitogenome (). Four PCGs, two rRNA genes and eight tRNA genes were encoded by the light strand, while others were located on the heavy strand.
Figure 2. The mitochondrial genome map of Peleteria iavana. Arrangement of 37 genes represented in the map, including 13 protein-coding genes, 22 tRNA genes, and two rRNA genes. A circular mitochondrial genome map was drawn using Geneious Prime 2020.2.2 (Kearse et al. Citation2012). 
 control region,
control region,  rRNA gene,
rRNA gene,  tRNA gene.
tRNA gene.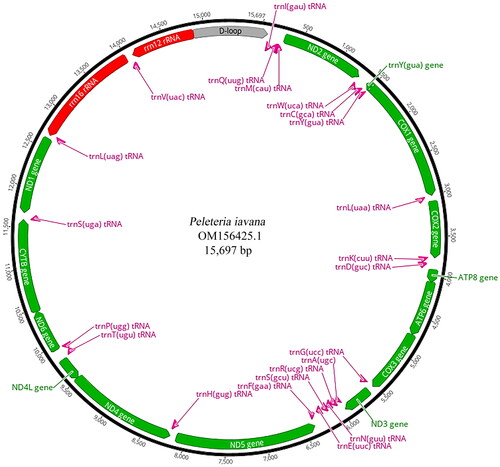
The 13 PCGs accounted for 71.1% of the complete mitogenome of P. iavana (11,160 bp). PCGs utilized a variety of start codons including the standard ATN, except for the nonstandard TCG (COI) and TTG (NDI). The most frequent start codon was ATG, which covered six PCGs (COII, ATP6, COIII, ND4, ND4L, CYTB). Five PCGs (ND2, ATP8, ND3, ND5, ND6) started with ATT codon. The stop codon TAA was assigned to most of the PCGs (ND2, ATP8, ATP6, COIII, ND3, ND4L, ND6 and ND1), but an incomplete stop codon T was used by four PCGs (COI, COII, ND5 and ND4), CYTB terminated with stop codon TAG. Besides, the length of 22 tRNAs in the mitogenome ranged from 63 bp (tRNAAla, tRNAArg, tRNACys) to 72 bp (tRNAVal).
As shown in , a phylogenetic analysis based on 13 PCGs revealed that P. iavana and (Janthinomyia sp.+Lydina aenea) clustered together. Additionally, Hamaxiella brunnescens is closely related to ((Cylindromyia+Dexiinae)+Macquartia spp.). Therefore, the subfamily Tachininae was not assigned to be a monophyletic group.
Conclusion
In the present study, we first reported the mitochondrial genome of P. iavana. It was a closed circular molecule of 15,697 bp in length, containing 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and a control region. The phylogenetic analysis suggested that Tachinidae was a monophyletic group and subfamily Tachininae has not formed a monophyly. P. iavana and (Janthinomyia sp.+Lydina aenea) clustered together in the basal position. Hamaxiella brunnescens of tribe Palpostomatini from subfamily Tachininae was the sister group of (Dexiinae + Cylindromyini), the result was consistent with the previous study on the phylogeny of Tachinidae based on the morphology and molecular data (Stireman et al. Citation2019). Eventually, as the sister group to Hamaxiella brunnescens, four species of genus Macquartia clustered together with high supporting values, the phylogenetic tree showed a similarity to the phylogenetic relationships of the Tachinidae in Li et al. (Li et al. Citation2022). These results provided an important basis for further studies on the mitochondrial genomics and phylogenetics of the family Tachinidae.
Ethical approval
Ethical approval (No. 1900074) was granted by the Animal Care Welfare Committee of Guizhou Medical University for the study.
Author contributions
According to International Committee of Medical Journal Editors (ICMJE) criteria, all authors including Peng Zhang, Yan Zhi, Chunchun Guo, Chuntian Zhang and Jiayu Liu participated in the conception and design of the work, acquisition and analysis of data, drafting and revising the work, and final approval of the version to be published. Also, all authors agreed to be accountable for all aspects of the work.
Supplemental Material
Download MS Word (179.4 KB)Supplemental Material
Download JPEG Image (993.6 KB)Supplemental Material
Download JPEG Image (2.4 MB)Supplemental Material
Download PNG Image (7.8 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. OM156425.1. The associated BioProject, SRA and Bio-Sample numbers are PRJNA800680, SRR18030147 and SAMN25276555, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Hou P, Ding SM, Li X, Yang D, Zhang CT, Wang Q. 2018. The mitochondrial genome of Drino sp. (Diptera, Tachinidae). Mitochondrial DNA B Resour. 3(2):886–887.
- Hou P, Gao S, Li X, Yang D, Zhang CT. 2019. The mitochondrial genome of Janthinomyia sp. (Diptera, Tachinidae). Mitochondrial DNA B Resour. 4(1):1601–1602.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Li HN, Zhang BH, Pei WY, Sun HR, Chen JL, Gao XZ, Peng HL, Zhang D, Zhang CT. 2022. Four new species of Macquartia (Diptera: Oestroidea) from China and phylogenetic implications of Tachinidae. Insects. 13(12):1096.
- Li X, Ding SM, Hou P, Liu XY, Zhang CT, Yang D. 2017. Mitochondrial genome analysis of Ectophasia rotundiventris (Diptera, Tachinidae). Mitochondrial DNA B Resour. 2(2):457–458.
- Luo Y, Zhi Y, Zhang CT, Yang M, Liu JY. 2021. Complete mitochondrial genome of Compsilura concinnata (Meigen) (Diptera, Tachinidae). Mitochondrial DNA B Resour. 6(3):905–906.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63–e63.
- Nelson LA, Lambkin CL, Batterham P, Wallman JF, Dowton M, Whiting MF, Yeates DK, Cameron SL. 2012. Beyond barcoding: a mitochondrial genomics approach to molecular phylogenetics and diagnostics of blowflies (Diptera: calliphoridae). Gene. 511(2):131–142.
- O’Hara JE, Henderson SJ. 2020. World genera of the Tachinidae (Diptera) and their regional occurrence. Version 11.0. PDF document, pp. 90. https://www.uoguelph.ca/nadsfly/Tach/WorldTachs/Genera/Gentach_ver11.pdf
- O’Hara JE, Henderson SJ, Wood DM. 2020. Preliminary checklist of the Tachinidae (Diptera) of the world. Version 2.1. PDF document, pp. 1039. https://www.uoguelph.ca/nadsfly/Tach/WorldTachs/Checklist/Tachchlist_ver2.1.pdf
- O’Hara JE, Shima H, Zhang CT. 2009. Annotated catalogue of the Tachinidae (Insecta: diptera) of China. Zootaxa. 2190(1):1–236.
- Pei WY, Yan LP, Yang N, Zhang CT, Zheng CY, Yang J, Zhang D. 2019. First report of mitogenome of Subclytia rotundiventris (Diptera, Tachinidae) yielded by next-generation sequencing. Mitochondrial DNA B Resour. 4(2):2910–2911.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33(Web Server):W686–W689.
- Seo BY, Cho J, Lee GS, Park J, Park J. 2019. The complete mitochondrial genome of Exorista japonica (Townsend, 1909) (Diptera: tachinidae). Mitochondrial DNA B Resour. 4(2):2244–2245.
- Shao YJ, Hu XQ, Peng GD, Wang RX, Gao RN, Lin C, Shen WD, Li R, Li B. 2012. Structure and evolution of the mitochondrial genome of Exorista sorbillans: the Tachinidae (Diptera: calyptratae) perspective. Mol Biol Rep. 39(12):11023–11030.
- Stireman JO, O'Hara JE, Wood DM. 2006. Tachinidae: evolution, behavior, and ecology. Annu Rev Entomol. 51:525–555.
- Stireman JO, Cerretti P, O'Hara JE, Blaschke JD, Moulton JK. 2019. Molecular phylogeny and evolution of world Tachinidae (Diptera). Mol Phylogenet Evol. 139:106358.
- Wang R, Zhi Y, Zhang CT, Yang M, Liu JY. 2022. Complete mitochondrial genome of Cylindromyia (Calocyptera) intermedia from Guiyang, China, and phylogeny of Phasiinae (Diptera: tachinidae). Mitochondrial DNA B Resour. 7(6):1008–1010.
- Wang R, Zhi Y, Yao QY, Zhang CT, Liu JY. 2022. Characterization of the complete mitochondrial genome of Gymnosoma dolycoridis (Diptera, Tachinidae) and phylogenetic analysis. Mitochondrial DNA B Resour. 7(8):1435–1437.
- Yan LP, Pei WY, Zhang CT, Zhang D. 2021. First report of the mitogenome of Hamaxiella brunnescens (Diptera, Tachinidae) from Beijing, China. Mitochondrial DNA B Resour. 6(3):862–864.
- Zhao Z, Su TJ, Chesters D, Wang SD, Ho SYW, Zhu CD, Chen XL, Zhang CT. 2013. The mitochondrial genome of Elodia flavipalpis Aldrich (Diptera: Tachinidae) and the evolutionary timescale of tachinid flies. PLoS One. 8(4):e61814.