Abstract
Trees of Engelhardia are important components of subtropical and tropical forests in South-eastern Asia with great ecological and economic values. However, phylogenetic relationships within Engelhardioideae (Juglandaceae) remains obscure. In this study, we report the first complete chloroplast genome sequences of Engelhardia sensu stricto, Engelhardia hainanensis Chen, a rare species endemic in southern China. Its complete chloroplast genome is 161,574 bp in length, with a typical quadripartite structure that includes a large single-copy region of 91,158 bp, a small single-copy region of 18,790 bp, and its GC content is 35.8%. A total of 128 genes were identified, including 83 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Furthermore, a phylogenetic tree of Juglandaceae was constructed based the complete chloroplast genome sequence, which strongly support the three-subfamily classification system in Juglandaceae, and E. hainanensis was resolved sister to two Alfaropsis species. This study provides valuable genomic information for the species identification and phylogenetic study of Juglandaceae.
Introduction
Engelhardia Leschenault ex Blume is a deciduous, semievergreen or evergreen tree group of Juglandaceae which distributes in South-Eastern Asia from eastern Pakistan to New Guinea (Lu et al. Citation1999). Members of this group are important component in the subtropical and tropical forests, whose leaves are used for drinking as tea while its bark are used to poison fish in local community. Contrary to great ecological and economic value, the subgeneric classification and species number of Engelhardia is open to question (Meng et al. Citation2022). Engelhardia is suggested divided into genus Engelhardia sensu stricto (s.s.) which contains ca. five species, and a monotypic genus Alfaropsis Iljinsk. which contains only Alfaropsis roxburghiana (Wall.) Iljinsk. (Iljinskaya Citation1993). A distinct morphological difference between Engelhardia s.s. and Alfaropsis lies in that prophyllum is obvious and envelops fruit in the former while it is absent in the latter. Engelhardia hainanensis Chen (Chen Citation1981) is a rare species which has the longest winged bracts out of fruit (ca. 8cm, ) in the genus, and it is thought endemic in Hainan Island, China. However, we discover it in Guangxi Zhuang Autonomous Region during our recent field investigation. In order to better understand the phylogenetic relationships within Engelhardia and Juglandaceae, and provide more genomic data for accurate identification of the toxic Engelhardia species, we sequenced and analyzed the first complete chloroplast genome of Engelhardia s.s., E. hainanensis.
Materials and methods
Sample collection and preservation
The fresh leaves of E. hainanensis were collected at Lingyun County in Guangxi, China (N24°31′52.86″, E106°27′43.67″). Leaves were dried with silica gel in the field. Once completely dried, the leaf tissues were stored at −20 °C freezer until further use. Voucher specimen (collector and collection number: Xian-Yun Mu ([email protected]) and MU4973) is deposited at the herbarium of Beijing Forestry University (www.bjfu.edu.cn).
DNA extraction and sequencing, and cleaning raw reads
Genomic DNA were extracted from the above leaf tissues using the DNAsecure Plant Kit (Tiangen Biotech Co. Ltd., Beijing, China), and sequenced by next-generation sequencing method on Illumina Hiseq X Ten platform. In total of 4.1 Gb of 150-bp clean reads were generated for chloroplast genome assembly.
Assembly, annotation, and visualization
Complete chloroplast genome assembly was performed on GetOrganelle 1.7.5 (Jin et al. Citation2020), the annotation was performed on CPGAVAS2 (Shi et al. Citation2019), and further verification by Geneious Prime 2022 (Kearse et al. Citation2012) with A. roxburghiana (Ling and Zhang Citation2020) as a reference. The complete plastome of E. hainanensis has been deposited in GenBank under accession number OM302449. A circular map of its plastome was visualized using the CPGView online web (http://www.1kmpg.cn/cpgview, Liu et al. Citation2023)
Phylogenetic reconstruction
To determine the phylogenetic relationships of E. hainanensis in Juglandaceae, 40 species in Juglandaceae and three species in Betulaceae, Fagaceae, and Myricaceae family were selected based on previous researches (e.g., Zhang et al. Citation2019; Mu et al. Citation2020; Song et al. Citation2020). These related plastome sequences were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/). All sequences were aligned using MAFFT (Katoh et al. Citation2019). A maximum likelihood tree with bootstrap value (1000 replicates) was constructed based on GTR + G model in RAxML software (Kozlov et al. Citation2019). The final tree was edited using the iTOL version 5.0 online web (https://itol.embl.de/) (Letunic and Bork Citation2016).
Results and discussion
Characterization of the chloroplast genome
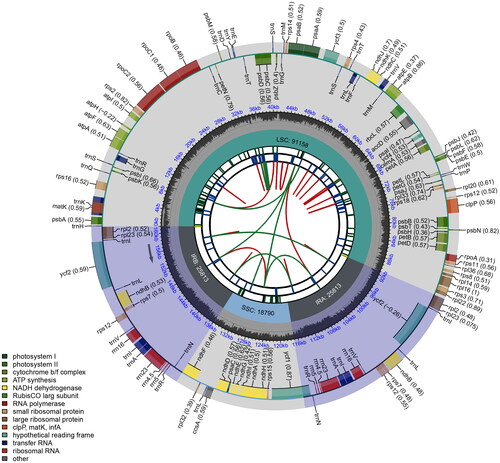
The complete chloroplast genome of E. hainanensis was assembled correctly with 161,574 bp in length (Figure S1), and its schematic circular map was showed in . The genome has a typical quadripartite structure, including a large single-copy (LSC) region of 91,158 bp, a small single-copy (SSC) region of 18,790 bp, and two inverted repeat (IRa and IRb) regions of 25,813 bp for each. Overall, the average GC content was 35.8%. A total of 128 genes (110 unique genes) were identified in the chloroplast genome of E. hainanensis, including 83 protein-coding genes (77 unique genes), 37 tRNA genes (29 unique genes), and 8 rRNA genes (4 unique genes). Among them, there were 12 protein-coding genes (rps16, atpF, rpoC1, ycf3, clpP, petB, petD, rpl16, rpl2, ndhB, ndhA, rps12, Figure S2) and 6 tRNA genes (trnK-UUU, trnG-UCC, trnL-UAA, trnV-UAC, trnI-GAU, trnA-UGC) with introns. The trans-splicing gene rps12 had three unique exons (Figure S3).
Figure 2. Schematic circular map of overall features of Engelhardia hainanensis chloroplast genome. Graphic showing features of its plastome was generated using CPGview. The map contains six tracks. From the inner circle, the first track depicts the dispersed repeats connected by red (forward direction) and green (reverse direction) arcs, respectively. The second track shows the long tandem repeats as short blue bars. The third track displays the short tandem repeats or microsatellite sequences as short bars with different colors. The fourth track depicts the sizes of the inverted repeats (IRa and IRb), small single-copy (SSC), and large single-copy (LSC). The fifth track plots the distribution of GC contents along the plastome. The sixth track displays the genes belonging to different functional groups with different colored boxes. The outer and inner genes are transcribed in the clockwise and counterclockwise directions, respectively.
Phylogenetic analysis of E. hainanensis within the family Juglandaceae
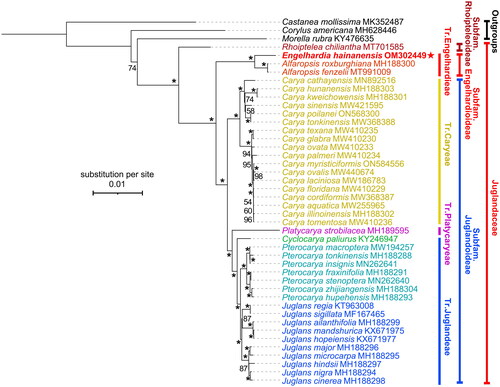
Phylogenetic relationships intra Juglandoideae are frequently investigated (Zhang et al. Citation2019; Mu et al. Citation2020; Song et al. Citation2020), while those within Engelhardioideae are obscure (Zhang et al. Citation2020). We reconstructed the phylogeny of Juglandaceae based on whole chloroplast genome sequences of 40 reported species and the new data in this study, and Castanea mollissima Blume (Blume Citation1851) (Fagaceae), Corylus americana Walter (Walter Citation1788) (Betulaceae), and Morella rubra Lour. (Loureiro Citation1790) (Myricaceae) were selected as outgroups. Three main clades are recovered with full support values, which corresponding to the three subfamilies of Juglandaceae, i.e., subfamily Rhoipteleoideae, subfamily Engelhardioideae, and subfamily Juglandoideae (). This is consistent with previous phylogenetic analyses (e.g., Zhang et al. Citation2019; Mu et al. Citation2020; Song et al. Citation2020). Further, E. hainanensis was resolved a sister clade to the two Alfaropsis species (i.e., A. roxburghiana and A. fenzelii). A large sampling scheme covering more species is needed for further clarification of phylogenetic relationships intra subfamily Engelhardioideae.
Figure 3. Phylogenetic tree of Juglandaceae inferred from maximum-likelihood (ML) method based on concatenated complete chloroplast genome sequence of 41 species with Corylus americana, Castanea mollissima, and Morella rubra as outgroup. *The clade support value of 100 that was generated from the maximum likelihood method. The red star represents the assembled plastome sequence (i.e., Engelhardia hainanensis OM302449) in this study. GenBank accession numbers of the following sequences were used: Castanea mollissima MK352487 (Zhu et al. Citation2019), Corylus americana MH628446 (Zhao et al. Citation2020), Morella rubra KY476635 (Liu et al. Citation2017), Rhoiptelea chiliantha MT701585 (Jin et al. Citation2020), Alfaropsis roxburghiana MH188300 (Zhou et al. Citation2021), Alfaropsis fenzelii MT991009 (Liu et al. Citation2021), Carya cathayensis MN892516 (Shen et al. Citation2022), Carya hunanensis MH188303 (Zhou et al. Citation2021), Carya kweichowensis MH188301 (Zhou et al. Citation2021), Carya sinensis MW421595 (Xi et al. Citation2022), Carya poilanei ON568300 (Xi et al. Citation2022), Carya tonkinensis MW368388 (Xi et al. Citation2022), Carya texana MW410235 (Xi et al. Citation2022), Carya glabra MW410230 (Xi et al. Citation2022), Carya ovata MW410233 (Xi et al. Citation2022), Carya palmeri MW410234 (Xi et al. Citation2022), Carya myristiciformis ON584556 (unpublished), Carya ovalis MW410234 (Xi et al. Citation2022), Carya laciniosa MW186783 (Zhai et al. Citation2021), Carya floridana MW410229 (Xi et al. Citation2022), Carya cordiformis MW368387 (Xi et al. Citation2022), Carya aquatica MW255965 (Xi et al. Citation2022), Carya illinoinensis MH188302 (Zhou et al. Citation2021), Carya tomentosa MW410236 (Xi et al. Citation2022), Platycarya strobilacea MH189595 (Zhou et al. Citation2021), Cyclocarya paliurus KY246947 (Hu et al. Citation2017), Pterocarya macroptera MW194257 (Yan et al. Citation2021), Pterocarya tonkinensis MH188288 (Zhou et al. Citation2021), Pterocarya insignis MN262641 (Mu et al. Citation2020), Pterocarya fraxinifolia MH188291 (Zhou et al. Citation2021), Pterocarya stenoptera MN262640 (Mu et al. Citation2020), Pterocarya zhijiangensis MH188304 (Zhou et al. Citation2021), Pterocarya hupehensis MH188293 (Zhou et al. Citation2021), Juglans regia KT963008 (Hu et al. Citation2017), Juglans sigillata MF167465 (Dong et al. Citation2017), Juglans ailanthifolia MH188299 (Zhou et al. Citation2021), Juglans mandshurica KX671975 (Hu et al. Citation2017), Juglans hopeiensis KX671977 (Hu et al. Citation2017), Juglans major MH188296 (Zhou et al. Citation2021), Juglans microcarpa MH188295 (Zhou et al. Citation2021), Juglans hindsii MH188297 (Zhou et al. Citation2021), Juglans nigra MH188294 (Zhou et al. Citation2021), Juglans cinerea MH188298 (Zhou et al. Citation2021).
Conclusions
We reported the first complete chloroplast genome sequences of Engelhardia s.s., E. hainanensis. The assembly circular plastome was 16,534 bp in length. The phylogenetic analysis results strongly supported the three-subfamily classification system in Juglandaceae, and E. hainanensis was a resolved sister to two Alfaropsis species. The plastome sequence of E. hainanensis presented here provides valuable genomic information for further species identification and phylogenetic study of Juglandaceae.
Ethical approval
This study includes no endangered plant samples, and the sampling site is not located in any protected area. The collection of plant materials is in accordance with local regulations and obtain the permission of local authorities.
Author contributions
XYM conceived and designed the work. XYM and YHQ performed field collection and identification of plant material. XYM and YMW assembled and annotated the plastome sequence. YMW prepared genome sequences, performed phylogenetic analysis, and visualized figures. XYM and YMW wrote the manuscript. YMW, XYM and YHQ reviewed drafts of the paper. All authors discussed the results, commented on the manuscript, and approved the current version of the manuscript.
Supplemental Material
Download TIFF Image (391 KB)Supplemental Material
Download TIFF Image (926.4 KB)Supplemental Material
Download TIFF Image (28.6 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. OM302449. The associated BioProject, SRA, and BioSample numbers are PRJNA797548, SRP355546 and SAMN25010399, respectively.
Additional information
Funding
References
- Blume CL. 1851. Museum botanicum Lugduno-Batavum, sive, Stirpium exoticarum novarum vel minus cognitarum ex vivis aut siccis brevis expositio et descriptio. Lugdunum Batavorum: Nabu Press; p. 286.
- Chen PY. 1981. Two new species of Engelhardia from Hainan Island. Acta Phytotaxon Sin. 19(2):250–252.
- Dong WP, Xu C, Li WQ, Xie XM, Lu YZ, Liu YL, Jin XB, Suo ZL. 2017. Phylogenetic resolution in Juglans based on complete chloroplast genomes and nuclear DNA sequences. Front Plant Sci. 8:1148.
- Hu YH, Woeste KE, Zhao P. 2017. Completion of the chloroplast genomes of five Chinese Juglans and their contribution to chloroplast phylogeny. Front Plant Sci. 7:1955.
- Hu YH, Yan J, Feng XJ, Dang M, Woeste KE, Zhao P. 2017. Characterization of the complete chloroplast genome of wheel wingnut (Cyclocarya paliurus), an endemic in China. Conserv Genet Resour. 9(2):273–275.
- Iljinskaya IA. 1993. Alfaropsis, a new genus of the Juglandaceae. Bot Zhurn. 78:79–83.
- Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Jin L, Zhou GH, He ML, Yang ZY, Yan HJ. 2020. The complete plastome and phylogenetic analysis of Rhoiptelea chiliantha (Juglandaceae). Mitochondrial DNA Part B. 5(3):3091–3092.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 35(21):4453–4455.
- Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44(W1):W242–W245.
- Ling LZ, Zhang SD. 2020. Characterization of the complete chloroplast genome of Engelhardia roxburghiana (Juglandaceae). Mitochondrial DNA Part B. 5:250–251.
- Liu LX, Li R, Worth JRP, Li X, Li P, Cameron KM, Fu CX. 2017. The complete chloroplast genome of Chinese bayberry (Morella rubra, Myricaceae): implications for understanding the evolution of Fagales. Front Plant Sci. 8:968.
- Liu M, Lu JS, Li Y, Zhang LS. 2021. Complete chloroplast genome of Engelhardtia fenzlii (Juglandaceae). Mitochondrial DNA Part B. 6(1):288–289.
- Liu SY, Ni Y, Li JL, Zhang XY, Yang HY, Chen HM, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704.
- Loureiro JD. 1790. Flora Cochinchinensis. Ulyssipo: Typis et expensis Academicis; p. 548.
- Lu AM, Stone DE, Grauke LJ. 1999. Juglandaceae. In Wu ZY, Raven PH, editors. Flora of China. Vol. 4. Beijing & St. Louis: Science Press & Missouri Botanical Garden Press; p. 277–285.
- Meng HH, Zhang CY, Low SL, Li L, Shen JY, Zhang Y, Huang PH, Zhou SS, Tan YH, Li J. 2022. Two new species from Sulawesi and Borneo facilitate phylogeny and taxonomic revision of Engelhardia (Juglandaceae). Plant Diversity. 44(6):552–564.
- Mu XY, Tong L, Sun M, Zhu YX, Wen J, Lin QW, Liu B. 2020. Phylogeny and divergence time estimation of the walnut family (Juglandaceae) based on nuclear RAD-Seq and chloroplast genome data. Mol Phylogenet Evol. 147:106802.
- Shen JS, Li XQ, Chen X, Huang XL, Jin SH. 2022. The complete chloroplast genome of Carya cathayensis and phylogenetic analysis. Genes. 13(2):369.
- Shi LC, Chen HM, Jiang M, Wang LQ, Wu X, Huang LF, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47:W65–W73.
- Song YG, Li Y, Meng HH, Fragnière Y, Ge BJ, Sakio H, Yousefzadeh H, Bétrisey S, Kozlowski G. 2020. Phylogeny, taxonomy, and biogeography of Pterocarya (Juglandaceae). Plants. 9:1524.
- Walter T. 1788. Flora Caroliniana. p. 236. London: John Fraser.
- Xi JW, Lv SB, Zhang WP, Zhang JB, Wang KT, Guo HB, Hu J, Yang Y, Wang JH, Xia GH, et al. 2022. Comparative plastomes of Carya species provide new insights into the plastomes evolution and maternal phylogeny of the genus. Front Plant Sci. 13:990064.
- Yan H, Zhang JS, Ye JF, Guo CC, Xiang XG. 2021. Characterization of the complete chloroplast genome of Pterocarya macroptera var. delavayi (Juglandaceae). Mitochondrial DNA Part B. 6(4):1344–1345.
- Zhai M, Jia XD, Zhang JY, Guo ZR, Xuan JP. 2021. Characterization of the complete chloroplast genome of Carya laciniosa (F. Michx.) G. Don (Juglandaceae). Mitochondrial DNA Part B. 6(3):1261–1262.
- Zhang CY, Ling Low S, Song YG, Nurainas KG, Van Do T, Li L, Zhou SS, Tan YH, Cao GL, et al. 2020. Shining a light on species delimitation in the tree genus Engelhardia Leschenault ex Blume (Juglandaceae). Mol Phylogenet Evol. 152:106918.
- Zhang BW, Xu LL, Li N, Yan PC, Jiang XH, Woeste KE, Lin K, Renner SS, Zhang DY, Bai WN. 2019. Phylogenomics reveals an ancient hybrid origin of the Persian walnut. Mol Biol Evol. 36(11):2451–2461.
- Zhao TT, Wang GX, Ma QH, Liang LS, Yang Z. 2020. Multilocus data reveal deep phylogenetic relationships and intercontinental biogeography of the Eurasian-North American genus Corylus (Betulaceae). Mol Phylogenet Evol. 142:106658.
- Zhou HJ, Hu YH, Ebrahimi A, Liu PL, Woeste K, Zhao P, Zhang SX. 2021. Whole genome based insights into the phylogeny and evolution of the Juglandaceae. BMC Ecol Evol. 21(1):1–16.
- Zhu CC, Shi FH, Wang M, Zhao YQ, Chen Y, Geng GM. 2019. The complete chloroplast genome of a variety of Castanea mollissima ‘Hongli’ (Fagaceae). Mitochondrial DNA Part B. 4(1):993–994.

