Abstract
In this study, we obtained the complete mitochondrial genome of Hypseleotris cyprinoides, which was 16520 bp in length. The mitogenome contained 37 genes, including the typical set of 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, and 2 Ribosomal RNA (rRNA) genes. A, C, G, and T distribution was 28.57%, 29.91%, 16.99%, and 24.53%, respectively. The length of the total protein-coding genes was 11441 bp, which accounts for 66.80% of the whole mitochondrial genome. The Maximum Likelihood (ML) phylogenetic analysis based on the concatenated nucleotide sequences of 13 PCGs showed that H.cyprinoides as a sister species to Hypseleotris klunzingeri was clustered in the family Hypseleotris. The discovery of the complete mitochondrial genome of H.cyprinoides would help to conduct in-depth research on Hypseleotris.
1. Introduction
Hypseleotris cyprinoides (Valenciennes, 1837) belongs to the genus of Hypseleotris of Eleotridae family, Gobioidei of Perciformes, which distributes in the estuary region of China, Japan, Southeast Asian countries in Southwest Pacific. H.cyprinoides have a strongly laterally compressed head and body, a small mouth not reaching the anterior border of the orbit, an elongated body cavity with several anal pterygiophores preceding the first vertebral hemal spine, and an ovoid blotch at the dorsal base of the pectoral fin (, This image was taken by author Xinhe Ruan). Although it is classified into the Eleotridae by morphological method, there is almost no analysis of the gene sequence of H.cyprinoides (Thacker and Unmack Citation2005). The Mitochondrial genome has been widely used for phylogenetic studies in recent years, and several new perspectives other than traditional morphological classification have been proposed (Miya et al. Citation2003). Therefore, to clarify its evolutionary status at the level of molecular, we determined the mitochondrial genome sequence of H. cyprinoides and analyzed its evolutionary characteristics, which will help us to improve the data at the molecular level and clarify the phylogenetic relationship and taxonomic status of H. cyprinoides in this study.
2. Materials and methods
2.1. Sample collection and preservation
The specimen of Hypseleotris cyprinoides was obtained from Pingtung City, Taiwan Province, China (N22°0854′, E120°7474′) in November 2021. This specimen is deposited in the Laboratory of Aquatic Economic Animal Germplasm Resources and breeding Engineering, South China Agricultural University, China (Xinhe Ruan, [email protected]), under voucher number CHT2150001.
2.2. DNA extraction and sequencing and phylogenetic analysis method
Total genomic DNA was extracted using a modified cetyltrimethylammonium bromide (CTAB) method and applied to 500-bp paired-end library construction using the NEBNext Ultra DNA Library Prep Kit for Illumina sequencing. Sequencing was carried out on the Illumina NovaSeq 6000 platform (BIOZERON Co., Ltd., Shanghai, China), and using a run configuration of 2 × 150 bp to generate approximately 5 Gb of data for each sample. Eventually, clean Data was spliced using SPAdes v3.14.1 software, the assembled sequence was reordered and oriented according to the reference mitochondrial genome (Zhang et al. Citation2000), thus generating the final assembled mitochondrial genomic sequence. The mitogenome was assembled from 6934 Mb raw reads, with mean depth of 400×. The GC content obtained therein was 40.63%. The mitochondrion genes were annotated using the online MITOS tool, using default parameters to predict protein-coding genes, transfer RNA (tRNA) genes, and ribosome RNA (rRNA) genes. The base composition was calculated and the phylogenetic tree was built using MEGA X software (Kumar et al. Citation2018).
3. Results and discussion
3.1. Characteristics of H. cyprinoides mitochondrial genome
The complete mitogenome of H. cyprinoides was 16,520 bp in length (GenBank accession number: OM971860) and contained the typical set of 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 Ribosomal RNA (rRNA) genes (). And the software CGView was used to map the mitochondrial genome (). Its composition is similar to that of typical vertebrates (Miya et al. Citation2001). The distribution of A, C, G, and T was 28.57%, 29.91%, 16.99%, and 24.53% respectively. The length of the total protein-coding genes was 11,441 bp, which accounts for 66.80% of the whole mitochondrial genome, and the base composition was 25.96% for A, 31.19% for C, 16.51% for G, and 26.34% for T. Most of the mitochondrial genes were encoded in the H chain, except for ND6 and 8 tRNA genes (Gln, Ala, Asn, Cys, Tyr, Ser, Glu, and Pro), which were encoded by the L chain. Most PCGs began with a start codon ATG except the COX1 gene, which initiated with GTG. This is similar to the mitochondrial DNA of the Eleotridae family (Zang et al. Citation2016; Meng et al. Citation2016). Ten PCGs terminated with a complete stop codon TAA or TAG, whereas the other three, including the COX2, NAD4, and COB genes, ended with others.
Figure 2. Image and mitochondrion genome map of Hypseleotris cyprinoides. CDS: Coding sequence; tRNA: Transfer RNA; rRNA: Ribosomal RNA.
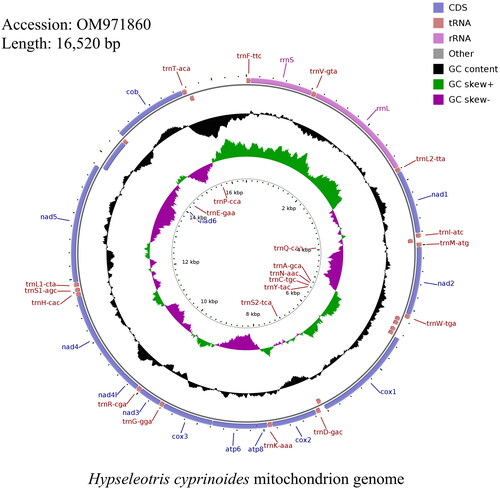
Table 1. The organization of the complete mitochondrial genome in Hypseleotris cyprinoides.
3.2. Phylogenetic analysis
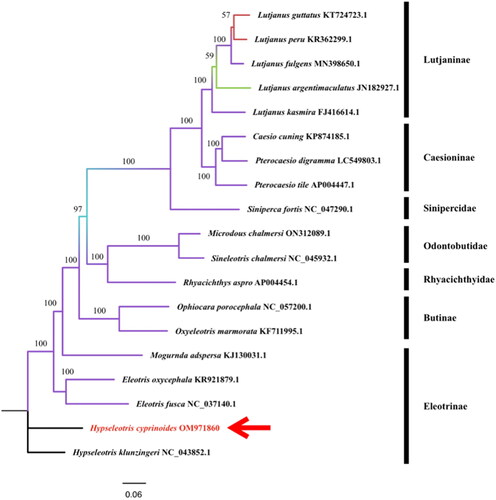
The Maximum Likelihood (ML) phylogenetic tree was built based on 13 PCG of 19 species’ complete mitochondrial genomes. All the mitochondrial gene sequences were downloaded from the NCBI gene bank. The number at each node has been obtained from the probability by 1000 bootstrap. From the phylogenetic tree (), we can see that the genome of H. cyprinoides is most similar to that of Hypseleotris klunzingeri among the species of Hypseleotris analyzed. This is consistent with the results of morphological classification. Both H. cyprinoides and Hypseleotris klunzingeri belong to the genus of Hypseleotris. According to Australian researchers (Schmidt and McDougall Citation2019), the Hypseleotris klunzingeri is a freshwater estuary fish endemic to Australia, which is comparable to where we collected the specimen. It is speculated that the separation of the Earth’s plates resulted in geographical isolation and the emergence of two comparable species. It will provide better phylogenetic insights into this species.
Figure 3. A phylogenetic tree for H. cyprinoides and other 18 species based on assembled nucleotide sequences of 13 protein-coding genes, two rRNA genes, and 22 tRNA genes. The base composition was calculated and the phylogenetic tree was built using MEGA X software. The number on each node indicates the values of the ultrafast bootstrap (UFB) of 1000 replications. The phylogenetic position of H. cyprinoides was marked with a red arrow. The following sequences were used: Eleotris oxycephala KR921879.1 (Meng et al. Citation2016), Eleotris fusca NC_037140.1 (unpublished), Hypseleotris klunzingeri NC 043852.1 (Schmidt et al. 2019b), Mogurnda adspersa KJ130031.1 (Perini et al. Citation2016), Ophiocara porocephala NC_057200.1 (unpublished), Oxyeleotris marmorata KF711995.1 (Xu et al. Citation2016), Microdous chalmersi ON312089.1 (Wang et al. Citation2019), Sineleotris chalmersi NC 045932.1 (Wang et al. Citation2019), Rhyacichthys aspro AP004454.1 (Miya et al. Citation2003), Lutjanus guttatus KT724723.1 (Bayona-Vásquez et al. Citation2017), Lutjanus peru KR362299.1 (Bayona-Vásquez et al. Citation2017), Lutjanus fulgens MN398650.1 (Afriyie et al. Citation2020), Lutjanus argentimaculatus JN182927.1 (unpublished), Lutjanus kasmira FJ416614.1 (unpublished), Casio cuing KP874185.1 (Zhan et al. Citation2017), Pterocaesio digramma LC549803.1 (Song et al. Citation2020), Pterocaesio tile AP004447.1 (Miya et al. Citation2003), Siniperca fortis NC 047290.1(Peng et al. Citation2020).
4. Conclusions
We reported the first complete mitochondrial genome assembly and annotation of H. cyprinoides using next-generation sequencing technology. The circular mitogenome was 16,520 bp in length, contained 37genes encoding 13 PCGs, 22 tRNAs, and two rRNAs. The phylogenetic tree was inferred by a Maximum-likelihood phylogenetic tree based on the sequences of 18 species, which supported that H. cyprinoides was grouped together with Hypseleotris klunzingeri. The mitochondrial genomic data of H. cyprinoides provided in this study will aid future research on the evolution, taxonomy, DNA barcoding, and population genetics of Hypseleotris species.
Ethical approval
This study was conducted with the guidelines of the Council of China and animal welfare requirements. Based on the recommendations of the Regulations for the Administration of Affairs Concerning Experimental Animals of China, the Institutional Animal Care and Use Committee of Guangdong Academy of Animal Science and Veterinary Medicine, South China Agricultural University approved all animal experiments.
Author contributions
Conceived and designed the experiments: Huihong Zhao and Chunli Zhang. Performed the experiments: Xinhe Ruan and Huitao Cheng. Analyzed the data and Wrote the paper: Zhongchao Pan. Final approval of the version to be published: Chunli Zhang. All authors have read and agreed to the published version of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under accession no. OM971860. The associated Bio-Project, SRA, and Bio-Sample numbers are PRJNA858347, SRR20183523, and SAMN29706239 respectively.
Additional information
Funding
References
- Afriyie G, Wang Z, Dong Z, Ayisi Larbi C, Asiedu B, Guo Y., 2020. Complete mitochondrial genome and assembled DNA barcoding analysis of Lutjanus fulgens (Valenciennes, 1830) and its comparison with other Lutjanus species. Ecol Evol. 10(15):7971–7980.
- Bayona-Vásquez NJ, Hernández-Álvarez CA, Glenn T, Domínguez-Domínguez O, Uribe-Alcocer M, Díaz-Jaimes P., 2017. Complete mitogenome sequences of the pacific red snapper (Lutjanus peru) and the spotted rose snapper (Lutjanus gutattus). Mitochondrial DNA Part A. 28(2):223–224.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K., 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Meng Y, Ma H, Ma C, Wei H, Liu Y, Zhang F, Wang W, Chen W, Zhao M, Chen F, et al. 2016. The complete mitochondrial genome of Eleotris oxycephala (Perciformes: Eleotridae). Mitochondrial DNA Part A. 27(5):3820–3821.
- Miya M, Kawaguchi A, Nishida M. 2001. Mitogenomic exploration of higher teleostean phylogenies: a case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol Biol Evol. 18(11):1993–2009.
- Miya M, Takeshima H, Endo H, Ishiguro NB, Inoue JG, Mukai T, Satoh TP, Yamaguchi M, Kawaguchi A, Mabuchi K, et al. 2003. Major patterns of higher teleostean phylogenies: a new perspective based on 100 complete mitochondrial DNA sequences. Mol Phylogenet Evol. 26(1):121–138.
- Peng M, Zhu W, Zeng D, Yang C, Chen X., 2020. The complete mitochondrial genome of Siniperca fortis (Perciformes: Sinipercidae). Mitochondrial DNA Part B. 5(2):1857–1858.
- Perini VDR, de Carvalho DC, Beheregaray LB, Prosdocimi F., 2016. The complete mitochondrial genome of the southern purple-spotted gudgeon Mogurnda adspersa (Perciformes: Eleotridae) through pyrosequencing. Mitochondrial DNA Part A. 27(1):380–382.
- Schmidt DJ, McDougall C. 2019. Complete mitogenomes of five ecologically diverse Australian freshwater fishes. Mitochondrial DNA Part B. 4(1):191–193.
- Song HY, Jung Y-H, Kim B, Choi YJ, Nguyen TV, Lee D-S., 2020. Complete mitochondrial genome of the double-lined fusileer, Pterocaesio digramma (Perciformes, Caesionidae): mitogenome characterization and phylogenetic analysis. Mitochondrial DNA Part B. 5(3):2617–2618.
- Thacker C, Unmack PJ. 2005. Phylogeny and biogeography of the eleotrid genus Hypseleotris (Teleostei: Gobioidei: Eleotridae), with redescription of H. cyprinoides. Rec Aust Mus. 57(1):1–13.
- Wang C, Zhang M, Cheng G, Chen X., 2019. The complete mitochondrial genome of Microdous chalmersi (Gobiiformes: Odontobutidae). Mitochondrial DNA Part B. 4(1):1979–1980.
- Xu Y, Hu Y, Bao B, Gong X., 2016. Complete mitochondrial DNA sequence of marble goby, Oxyeleotris marmorata (Bleeker, 1852). Mitochondrial DNA Part A. 27(2):817–818.
- Zang X, Yin D, Wang R, Yin S, Tao P, Chen J, Zhang G., 2016. Complete mitochondrial DNA sequence and phylogenic analysis of Oxyeleotris lineolatus (Perciformes, Eleotridae). Mitochondrial DNA Part A. 27(4):2414–2416.
- Zhan W, Chen R-Y, Shen K-N, Xu D-D, Hsiao C-D, Lou B., 2017. Next-generation sequencing yields the complete mitochondrial genome of the Redbelly yellowtail fusilier, Caesio cuning (Teleostei: Caesionidae). Mitochondrial DNA Part A. 28(1):125–126.
- Zhang Z, Schwartz S, Wagner L, Miller W., 2000. A greedy algorithm for aligning DNA sequences. J Comput Biol. 7(1–2):203–214.

