Abstract
The complete mitogenome sequence of Eothenomys eleusis Thomas 1911 was determined using PCR. A circular double-stranded structure makes up the mitochondrial genome of E. eleusis. The complete length of the mitochondrial genome is 16,419 bp. The mitochondrial genome of E. eleusis included 13 protein-coding genes, 1 control region, 22 tRNA genes, 2 rRNA genes and 1 origin of L strand replication. The total base composition of E. eleusis mitochondrial genome was A (32.6%), T (26.3%), G (13.6%) and C (27.5%). We found significant A-T skew in base composition, especially in control regions and protein-coding genes. E. eleusis was supported by bootstrap values of 100%. This study verifies the evolutionary status of E. eleusis in Myodini tribe of Cricetidae at the molecular level. The mitochondrial genome would be a significant supplement for the E. eleusis genetic background.
Introduction
Eothenomys eleusis Thomas 1911 belongs to the Rodentia order, Cricetidae family, Arvicolinae subfamily, Myodini tribe, genus Ethenomys (Wilson and Reeder et al. 2005). The species of the genus Ethenomys is relatively abundant in China (Liu et al. Citation2019). The genus is currently thought to have 17 species in China (Wei et al. Citation2021). E. eleusis is endemic to China (Wei et al. Citation2021). E. eleusis is widely distributed in south China (Liu et al. Citation2019). The taxonomy and phylogeny of Myodini tribe, especially in Eothenomys, have been controversial. In this study, the complete mitochondrial genome of E. eleusis was sequenced, and the phylogenetic relationships within Myodini tribe were analyzed.
Material & methods
A dead female E. eleusis was collected from Anshun City (26°21′48″N, 105°55′35″E), in Guizhou Province, China, in August 2021. This study got permission from the Laboratory Animal Welfare Ethics Committee of Mudanjiang Normal University. We described and measured the external morphology and skull morphology of the specimen (). We distinguished species using the key of Myodini in China (Tang et al. Citation2021). The Cyt b gene sequence of the specimen was blast in Genbank. The sequence (HM165380) the most similar to the specimen is from E. eleusis. The sample was stored at −75 °C before being used. The specimen was deposited at the Animal and Plant Herbarium of Mudanjiang Normal University (URL, Liu zhu and [email protected]) under the voucher number KQRS2021004. Genomic DNA was extracted from leg muscle using the EasyPure genomic DNA kit (TransGen Biotech Co., Beijing, China). Based on the reported mitochondrial genome of Eothenomys, we designed 15 pairs of primers for PCR. PCR gel images of 15 pairs of primers were taken (Figure S1). The first-generation sequencing technology was used in the sequencing of this study (the ABI 3730 sequencer, Boshi Biotechnology Co. Ltd., Haerbin, China). The sequences were assembled using DNA star, and analyzed and adjusted manually. The annotation of the E. eleusis mitochondrial genome was fulfilled using web-based services MITOS (http://mitos.bioinf.uni-leipzig.de/help.py) and software PhyloSuite v 1.2.2 (Zhang et al. Citation2020). The circular mitochondrial genome map of E. eleusis was drawn using OGDRAW 1.3.1 (Greiner et al. Citation2019). In this study, the molecular phylogeny of E. eleusis was investigated using the complete mitochondrial genomes of 14 species from 5 genera in Myodini tribe deposited in the GenBank. The phylogenetic tree was constructed by the nucleotide sequences of the complete mitochondrial genome without partitioning, through MEGA 11.0 software (Tamura et al. Citation2011). The phylogenetic tree was constructed using the Kimura 2-parameter model of Maximum Likelihood method with 1000 bootstrap replications.
Figure 1. The picture of external morphology and skull morphology. a: External morphology on the back, b: External morphology on the belly, c: Dorsal view of the maxilla, d: Ventral view of the maxilla. The morphological characters external morphology and skull morphology: Head and body length is 90 mm, Tail length is 42 mm, Tail length is about 45% of Head and body length, the color of back the body is black-brown, the molars have on root, the chewing surface of molars is wide. The specimen was collected by Liu zhu from the Anshun City (26°21′48″N, 105°55′35″E), in Guizhou Province, China, in August 2021. This picture was taken by Liu zhu.

Results
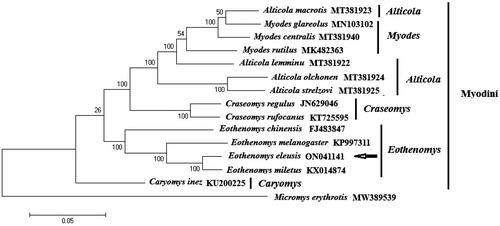
A circular double-stranded structure makes up the mitochondrial genome of E. eleusis (). The complete length of the mitochondrial genome is 16,419 bp. The mitochondrial genome of E. eleusis included 13 protein-coding genes, 1 control region, 22 tRNA genes, 2 rRNA genes and 1 origin of L strand replication (). The total base composition of E. eleusis mitochondrial genome was A (32.6%), T (26.3%), G (13.6%) and C (27.5%). We found significant A-T skew in base composition, especially in control regions and protein-coding genes. The ND6 gene and 8 tRNA genes of E. eleusis were encoded on the L strand. The other mitochondrial genes were encoded on the H strand (). GenBank received the annotated mitochondrial genome sequences, and the accession number ON041141 was given. Between the tRNA-Pro and tRNA-Phe genes is the control region of the mitochondrial genome (). The control region has no structural genes, but has only promoters and regulatory sequences for replication and transcription. The total length of 13 protein-coding genes sequences is 11,395 bp. The start codon for most protein-coding genes is ATG. The start codons for ND1, ND2 and ND3 is ATA or ATT. The stop codon for nine protein-coding genes is TAA, while ND4, COX3 use the incomplete stop codons (T– –). The other stop codons are TAG. The length of 22 tRNA genes are between 59 and 75 bp. The length of L-strand replication origin (OL) was 30 bp. On phylogenetic tree (), the 4 genera (Craseomys, Caryomys, Myopus and Eothenomys) in Myodini tribe formed independent branches. Alticola macrotis and A. lemminus of Alticola genus clustered on a branch with the Myopus genus (). E. eleusis was supported by bootstrap values of 100% (). Our results show that E. eleusis and E. miletus have a closer phylogenetic relationship ().
Figure 2. The circular mitochondrial genome map of E. Eleusis. The circular inside is L strand, and the circular outside is H strand. Yellow: NADH gene, pink: COX gene, green: ATP gene, purple: other genes, blue: tRNA, red: rRNA, nude: origin of L strand replication and control region.
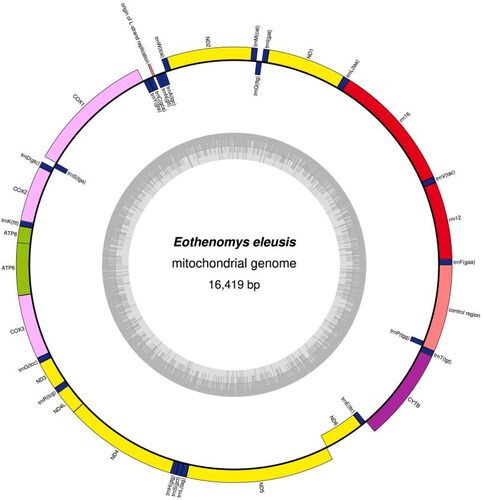
Figure 3. The phylogenetic tree constructed by the nucleotide sequences of complete mitochondrial genome without partitioning, through MEGA 11.0 software. The phylogenetic tree was constructed using the Kimura 2-parameter model of Maximum Likelihood method with 1000 bootstrap replications. Alticola strelzovi (Abramson et al. 2021), Alticola olchonensis (Abramson et al. 2021), Alticola lemminus (Abramson et al. 2021), Alticola macrotis (Abramson et al. 2021), Eothenomys miletus (Mu et al. 2019), Eothenomys melanogaster (Chen et al. 2016), Eothenomys chinensis (Yang et al. 2012), Eothenomys eleusis, Craseomys regulus (Abramson et al. 2021), Craseomys rufocanus (Lu et al. 2017), Myodes rutilus (Jin et al. 2019), Myodes centralis (Abramson et al. 2021), Myodes glareolus (Markováet et al. 2020), Caryomys inez (Yu et al. Citation2016). The out group is Micromys erythrotis (Cai et al. 2021).
Discussion & conclusions
The arrangement of genes in E. eleusis mitochondrial genome is consistent with other Cricetidae species (Chen et al. Citation2016; Cong et al. Citation2016; Luo and Liao Citation2016; Zhang et al. Citation2016; Jiang et al. Citation2018; Abramson et al. Citation2021; Baca et al. Citation2021). The phylogenetic relationship of this study is basically consistent with that of the recently published literature on Arvicolinae subfamilies (Liu et al. Citation2019; Abramson et al. Citation2021). This study verifies the evolutionary status of E. eleusis in Myodini tribe of Cricetidae at the molecular level. The mitochondrial genome would be a significant supplement for the E. eleusis genetic background.
Authors’ contributions
LIU Zhu, BI Zhong-Dan and ZHAO Jing-Yu collected the specimen and wrote the manuscript, BI Zhong-Dan, ZHAO Xin-Xu, QIAN Hong-An, MEI Xiu-Feng designed the experiment, identified the specimens and analyzed the data; CHEN Huan, ZHANG Jun-Sheng and PIAO Zhong-Wan were involved in the conception and design, funding acquisition and interpretation of the data. All authors agreed the final version to be published and agree to be accountable for all aspects of the work.
Supplemental Material
Download MS Word (302 KB)Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/, reference number ON041141.
Additional information
Funding
References
- Abramson NI, Bodrov SY, Bondareva OV, Genelt-Yanovskiy EA, Petrova TV. 2021. A mitochondrial genome phylogeny of voles and lemmings (Rodentia: Arvicolinae): evolutionary and taxonomic implications. PLOS One. 16(11):e0248198.
- Baca M, Popović D, Lemanik A, Fewlass H, Talamo S, Zima J, Ridush B, Popov V, Nadachowski A. 2021. The Tien Shan vole (Microtus ilaeus; Rodentia: Cricetidae) as a new species in the Late Pleistocene of Europe. Ecol Evol. 11(22):16113–16125.
- Cai H, Wang QQ, Zhao XX, Yao QQ, Wu N, Zhang JS, Liu Z. 2021. Sequencing and analysis of the complete mitochondrial genome of Micromys erythrotis from China and its phylogenetic analysis. Mitochondrial DNA Part B Resour. 6(5):1617–1620.
- Chen S, Chen G, Wei H, Wang Q. 2016. Complete mitochondrial genome of the Père David’s Vole, Eothenomys melanogaster (Rodentia: Arvicolinae). Mitochondrial DNA Part A DNA Mapp Seq Anal. 27(4):2496–2497.
- Cong H, Kong L, Liu Z, Wu Y, Motokawa M, Harada M, Li Y. 2016. Complete mitochondrial genome of the mandarin vole Lasiopodomys mandarinus (Rodentia: Cricetidae). Mitochondrial DNA Part A DNA Mapp Seq Anal. 27(1):760–761.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47:59–64.
- Jiang JQ, Wu SX, Chen JJ, Liu CZ. 2018. Characterization of the complete mitochondrial genome of short-tailed field vole, Microtus agrestis. Mitochondrial DNA Part B Resour. 3(2):845–846.
- Jin ZM, Yu CW, Liu Z. 2019. Sequencing and analysis of the complete mitochondrial genome of the northern red-backed vole (Myodes rutilus) from China. Mitochondrial DNA Part B. 4(1):1575–1577.
- Liu SY, Chen SD, He K, Tang M, Liu Y, Jin W, Li S, Li Q, Zeng T, Sun ZY, et al. 2019. Molecular phylogeny and taxonomy of subgenus Eothenomys (Cricetidae: Arvicolinae: Eothenomys) with the description of four new species from Sichuan, China. Zool J Linn Soc. 186(2):569–598.
- Lu T, Zhu M, Yi C, Si C, Yang C, Chen H. 2017. Complete mitochondrial genome of the gray red-backed vole (Myodes rufocanus) and a complete estimate of the phylogenetic relationships in Cricetidae. Mitochondrial DNA Part A DNA Mapp Seq Anal. 28(1):62–64.
- Luo G, Liao J. 2016. The complete mitochondrial genome of Allocricetulus eversmanni (Rodentia: Cricetidae). Mitochondrial DNA Part A DNA Mapp Seq Anal. 27(5):3102–3104.
- Marková S, Horníková M, Lanier HC, Henttonen H, Searle JB, Weider LJ, Kotlík P. 2020. High genomic diversity in the bank vole at the northern apex of a range expansion: the role of multiple colonizations and end‐glacial refugia. Mol Ecol. 29(9):1730–1744.
- Mu Y, Duan Y, Di Z, Wang Z, Wan LZ. 2019. The complete mitochondrial genome of the Yunnan red-backed vole Eothenomys miletus (Rodentia: Cricetidae) and its phylogeny. Mitochondrial DNA Part B. 4(1):1424–1425.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28(10):2731–2739.
- Tang MK, Chen ZH, Wang X, Chen ZX, He ZQ, Liu SY. 2021. A summary of phylogenetic systematics studies of Myodini in China (Rodentia: Cricetidae: Arvicolinae). Acta Theriologica Sinica. 41(1):71–81.
- Wei F, Yang Q, Wu Y, Jiang XL, Liu SY, Li BG, Yang G, Li M, Zhou J, Li S, et al. 2021. Catalogue of mammals in China (2021). Acta Theriologica Sinica. 41(5):487–501.
- Wilson DE, Reeder DM. 2005. Mammal Species of the World (3rd edition). Baltimore: The Johns Hopkins University Press.
- Yang C, Hao H, Liu S, Liu Y, Yue B, Zhang X. 2012. Complete mitochondrial genome of the Chinese oriental vole Eothenomys chinensis (Rodentia: Arvicolinae). Mitochondrial DNA. 23(2):131–133.
- Yu P, Kong L, Li Y, Cong H, Li Y. 2016. Analysis of complete mitochondrial genome and its application to phylogeny of Caryomys inez (Rodentia: Cricetidae: Arvicolinae). Mitochondrial DNA Part B, 1(1): 343–344.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.
- Zhang Z, Sun T, Kang C, Liu Y, Liu S, Yue B, Zeng T. 2016. The complete mitochondrial genome of lesser long-tailed Hamster Cricetulus longicaudatus (Milne-Edwards, 1867) and phylogenetic implications. Mitochondrial DNA Part A. 27(2):1303–1304.
