Abstract
Primula vialii Delavay ex Franch. (1905) is an alpine species with an ornamental value. In this study, we sequenced, assembled, and annotated the chloroplast genome of P. vialii. The results showed that it was a double-stranded, closed circular DNA with 154,897 bp in length, comprising a small single-copy (SSC) region of 17,766 bp, a large single-copy (LSC) region of 85,379 bp and a pair of inverted repeat (IR) regions of 25,876 bp. A total of 113 unique genes were annotated, including 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. The phylogenetic analysis revealed that P. vialii is closely related to Primula flaccida. The cp genomic data will be useful for systematics and evolutionary studies of Primula.
Introduction
Primula vialii Delavay ex Franch. (1905) is a perennial herb in the genus Primula (Primulaceae), which widely distributed in wet meadows and valleys at an altitude of 2800–4000 m in Northwest Yunnan and Southwest Sichuan of China (Hu and Kelso Citation1996). With its blue and red spires of flowers (), this extraordinary species presents an unique feature in Primula (Richards Citation2003). Because of its dark green basal leaves, it is invariably used as an ornamental foliage plant during vegetative growth period. However, previous studies have focused on tissue culture and rapid propagation (Li et al. Citation2005) of P. vialii rather than systematics and evolutionary studies. Here, we have sequenced and analyzed the chloroplast (cp hereafter) genome of P. vialii, which contributes to provide more cp genome genetic information further evolutionary research and phylogenetic study of Primula.
Materials and methods
Fresh leaves of an individual P. vialii were collected from the Alpine Botanical Garden of Lijiang City, Yunnan Province, China (26.997°N, 100.201°E). The voucher specimen (accession No. HY-12) was deposited at the Herbarium of Yunnan Normal University (Kunming, China; Jianlin Hang, [email protected]). Total genomic DNA was extracted from the fresh leaves using the modified CTAB method (Porebski et al. Citation1997) and stored in Huang’s Lab of Yunnan Normal University. The total DNA was fragmented into 300 bp short sequences following the manufacture’s protocoal (Illumina Inc., USA) to construct a DNA library, then sequenced on the Illumina Hiseq X Ten sequencing platform. We obtained 18,438,252 filtered reads of paired-end sequences (2 × 150 bp) and assembled the complete cp genome using NOVOPlasty v4.3.1 (Dierckxsens et al. Citation2017). To verify the accuracy of the assembly, we further mapped our clean reads back to the assembled cp genome to assess the depth of coverage (Figure S1). Taking the cp genome of Primula flaccida (Genbank accession No. NC_053595) as the reference sequence, the assembled genome was annotated using Geneious v2020.1.1 software (Kearse et al. Citation2012). The annotated cp genome sequences of P. vialii were deposited in the GenBank database under accession No. ON584545. The genome map and cis/trans-splicing genes map (Figure S2) was drawn using the CPGView program (http://www.1kmpg.cn/cpgview).
In order to investigate the phylogenetic relationship of P. vialii in the genus Primula, we downloaded 39 Primula cp genomes and 5 Primulaceae cp genomes (Glaux maritima (Liu et al. Citation2021), two Lysimachia species and two Androsace species) as outgroups from NCBI. The 45 cp genome sequences were aligned by software MAFFT v7.47 (Katoh and Standley Citation2013), then constructed a maximum likelihood tree using the software IQ-TREE 2 (Katoh and Standley Citation2013; Minh et al. Citation2020) under TVM + F + R3 best-fit model according to Bayesian information criterion (Kalyaanamoorthy et al. Citation2017). Branch supports were tested using ultrafast bootstrap (UFBoot) (Hoanget al. Citation2018) and SH-like approximate likelihood ratio test (SH-aLRT) (Guindon et al. Citation2010) with 10,000 replicates.
Results
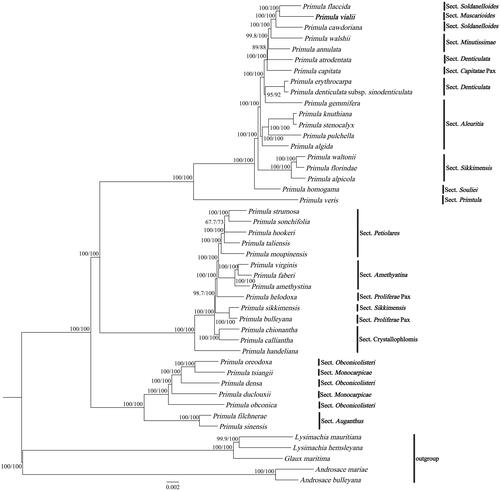
The cp genome of P. vialii was assembled as a double-stranded, closed-circular DNA molecule of 154,897 bp in length (), with an average coverage of 864.9 and GC contents of 36.9%. It has a typical quadripartite structure and comprises a large single-copy (LSC) region of 85,379 bp and a small single-copy (SSC) region of 17,766 bp, separated by two inverted repeat (IR) regions of 25,876 bp each. The GC contents of these four regions are 34.8%, 30.3%, 42.6%, 42.6%, respectively. In total, the cp genome has 132 genes, including 87 protein-coding genes, 37 tRNA genes, and 8 rRNA genes, of which 113 genes, 79 protein-coding genes, 30 tRNA genes, 4 rRNA genes are unique, respectively. Most of these genes are single-copy genes, while 8 protein-coding genes, 7 tRNA genes, and 4 rRNA genes were duplicated in the IR regions. The phylogentic tree () indicated the relationship between Primula, Androsace, Lysimachia, and Glaux in Primulaceae, and 40 Primula species formed a monophyletic clade. In addition, P. vialii is closely related to P. flaccida and P. cawdoriana in Sect. Soldanelloides.
Figure 2. Genomic map of Primula vialii chloroplast genome generated by CPGview (http://www.1kmpg.cn/cpgview/). The species name is shown in the left top corner. The map contains six tracks in default. From the center outward, the first track shows the dispersed repeats. The dispersed repeats consist of direct and Palindromic repeats, connected with red and green arcs. The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors. The small single-copy (SSC), inverted repeat (IRA and IRB), and large single-copy (LSC) regions are shown on the fourth track. The GC content along the genome is plotted on the fifth track. The base frequency at each site along the genome will be shown between the fourth and fifth tracks. The genes are shown on the sixth track. The optional codon usage bias is displayed in the parenthesis after the gene name. Genes belonging to different functional groups are color-coded. Genes drawn inside the circle are transcribed clockwise, and those outside are transcribed counterclockwise.
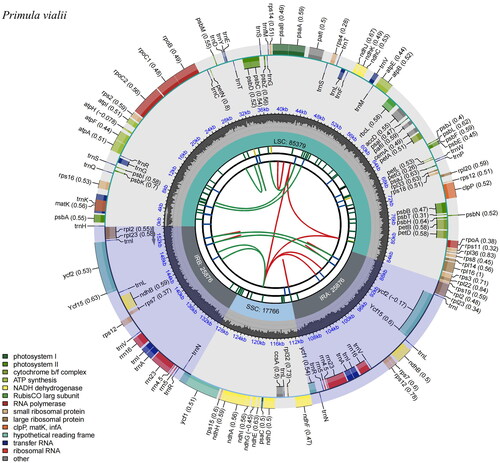
Figure 3. ML phylogenetic tree of Primula vialii and 44 Primulaceae species based on complete chloroplast genomes, the branch supports values were reported as SH-aLRT/UFBoot. The bolded font represents the chloroplast genome of P. vialii in this study. The following sequences were used: Primula flaccida NC_053595 (Wang et al. Citation2021), Primula cawdoriana NC_053586 (Wang et al. Citation2021), Primula walshii NC_053597 (Wang et al. Citation2021), Primula annulata NC_053608 (Wang et al. Citation2021), Primula atrodentata NC_053587 (Wang et al. Citation2021), Primula capitata NC_053589 (Wang et al. Citation2021), Primula erythrocarpa NC_053598 (Wang et al. Citation2021), Primula denticulata subsp. sinodenticulata NC_050247, Primula gemmifera NC_053590 (Wang et al. Citation2021), Primula knuthiana NC_039350 (Ren et al. Citation2018); Primula stenocalyx NC_058249 (Guo et al. Citation2021b), Primula pulchella NC_050246, Primula algida NC_053582 (Wang et al. Citation2021), Primula waltonii NC_058808, Primula florindae NC_053579 (Wang et al. Citation2021), Primula alpicola NC_053588 (Wang et al. Citation2021), Primula homogama NC_054305 (Sun et al. Citation2021), Primula veris NC_031428, Primula strumosa NC_053599 (Wang et al. Citation2021), Primula sonchifolia NC_053594 (Wang et al. Citation2021), Primula hookeri NC_053593 (Wang et al. Citation2021), Primula taliensis NC_053601 (Wang et al. Citation2021), Primula moupinensis NC_050244, Primula virginis NC_053581 (Wang et al. Citation2021), Primula faberi NC_053576 (Wang et al. Citation2021), Primula amethystina NC_053577 (Wang et al. Citation2021), Primula helodoxa NC_046771 (Zhang, Chen, et al. Citation2019), Primula sikkimensis NC_050243, Primula bulleyana NC_046947 (Chen, Zhang, et al. Citation2019), Primula chionantha NC_053583, (Wang et al. Citation2021), Primula calliantha MZ054238 (Yang et al. Citation2021), Primula handeliana NC_039348 (Ren et al. Citation2018), Primula oreodoxa NC_050848, Primula tsiangii NC_046755 (Chen, Yan, et al. Citation2019), Primula densa NC_058262 (Zhong et al. Citation2019), Primula duclouxii NC_058263 (Zhong et al. Citation2019), Primula obconica NC_046415 (Zhang, Yuan, et al. Citation2019), Primula filchnerae NC_051972 (Lu et al. Citation2020), Primula sinensis NC_030609 (Liu et al. Citation2016), Lysimachia mauritiana NC_060700 (Lee et al. Citation2022), Lysimachia hemsleyana NC_052863 (Ying et al. Citation2019), Glaux maritima NC_059901 (Liu et al. Citation2021), Androsace mariae NC_051991 (Guo et al. Citation2021a), Androsace bulleyana (NC_034641).
Discussion and conclusion
Within Primula clade, P. vialii, P. flaccida, and P. cawdoriana formed a monophyletic clade and were sister to each other (), which is consistent to previous studies based on noncoding cp DNA sequence (Mast et al. Citation2001). Mast (Citation2001) concluded that P. vialii belongs to Sect. Muscarioides, and members of Sect. Muscarioides are interdigitated with members of Sect. Soldanelloides. Although we reported the first complete cp genome of Sect. Muscarioides in this study, other complete cp genomes of Sect. Muscarioides are not sequenced yet. Therefore more phylogenetic researches are needed to exactly confirm the relationship of Sect. Muscarioides and Sect. Soldanelloides. Overall, the complete cp genome of P. vialii can be subsequently used for phylogenetic, taxonomic, and evolutionary studies of Sect. Muscarioides.
Ethics statement
This study does not involve ethical issues. The sample of this study is not a protected plant, and the sample collection has been approved by Lijiang Alpine Botanical Garden.
Authors contributions
Yunqi Liu performed the design of experiments, writing the original draft, and data curation. Shubao Wang, Li Zhang, and Rui Li performed the collection of P. vialii form wild fields, species identification, DNA extraction, and data analysis. Yuan Huang prepared the resources, supervised the project, made revisions to the manuscript, and final approval of the version to be published.
Supplemental Material
Download JPEG Image (357.4 KB)Supplemental Material
Download PNG Image (24.8 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov/] under accession no. ON584545. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA841839, SRR19391427, and SAMN28626130, respectively.
Additional information
Funding
References
- Chen S, Yan X, Hao G, Xu Y. 2019. The complete chloroplast genome of Primula tsiangii W. W. Smith (Primulaceae): a karst endemic primrose in Southwest China. Mitochondrial DNA B Resour. 4(2):2627–2628.
- Chen X, Zhang L, Li W, Huang Y, Wu Z. 2019. The complete chloroplast genome of Primula bulleyana, a popular ornamental species. Mitochondrial DNA B Resour. 4(2):3673–3674.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321.
- Guo Y, Ma L, Li J. 2021b. The complete chloroplast genome and phylogenetic analysis of Primula stenocalyx Maxim. Mitochondrial DNA B Resour. 6(11):3231–3232.
- Guo Y, Ma R, Xie J, Liu Q, Zhu Y, Yang W, Gao L, Ma Y. 2021a. The complete chloroplast genome of Androsace mariae. Mitochondrial DNA B Resour. 6(2):376–377.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522.
- Hu Q, Kelso S. 1996. Primulaceae. In: Wu CY, Raven PH, editors. Flora of China. Vol. 15. Beijing and St. Louis: Science Press and Missouri Botanical Garden. p. 122–123.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Lee Y, Yun N, Kang J, Choi S, Paik JH. 2022. The complete chloroplast genome of the medicinal plant Lysimachia mauritiana (Lamarck, 1792). Mitochondrial DNA B Resour. 7(3):554–555.
- Li Y, Xie WJ, He WJ, He JW. 2005. Tissue culture and rapid propagation of Primula vialii Franch. Plant Physiol Commun. 41(6):796.
- Liu YQ, Chen X, Zhang L, Huang Y. 2021. The complete chloroplast genome of Glaux maritima, a monotypic species. Mitochondrial DNA B Resour. 6(8):2137–2138.
- Liu TJ, Zhang CY, Yan HF, Zhang L, Ge XJ, Hao G. 2016. Complete plastid genome sequence of Primula sinensis (Primulaceae): structure comparison, sequence variation and evidence for accD transfer to nucleus. PEER J. 4:e2101.
- Lu Y, Liu PL, Sun YF, Li SF. 2020. The complete chloroplast genome sequence of Primula filchnerae Knuth (Primulaceae), an endangered species in China. Mitochondrial DNA B Resour. 5(3):2047–2048.
- Mast AR, Kelso S, Richards AJ, Lang DJ, Feller DM, Conti E. 2001. Phylogenetic relationships in Primula L. and related genera (Primulaceae) based on noncoding chloroplast DNA. Int J Plant Sci. 162(6):1381–1400.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Porebski S, Bailey LG, Baum BR. 1997. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep. 15(1):8–15.
- Ren T, Yang Y, Zhou T, Liu ZL. 2018. Comparative plastid genomes of Primula species: sequence divergence and phylogenetic relationships. IJMS. 19(4):1050.
- Richards J. 2003. Primula. 2nd ed. London: Batsford. p. 158–163.
- Sun HY, Zhong L, Guo YJ, Zhou W, Wu ZK. 2021. The complete chloroplast genome of a distylous-homostylous species, Primula homogama (Primulaceae). Mitochondrial DNA B Resour. 6(2):393–394.
- Wang XJ, Barrett SCH, Zhong L, Wu ZK, Li DZ, Wang H, Zhou W. 2021. The genomic selfing syndrome accompanies the evolutionary breakdown of heterostyly. Mol Biol Evol. 38(1):168–180.
- Yang X, Huang Y, Li Z, Chen J. 2021. Complete plastid genome of Primula calliantha Franch. (Primulaceae): an alpine ornamental plant endemic to Hengduan Mountain, China. Mitochondrial DNA B Resour. 6(9):2643–2645.
- Ying Z, Wang Q, Yu S, Liao G, Ge Y, Cheng R. 2019. The complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Lysimachia hemsleyana. Mitochondrial DNA B Resour. 4(2):3878–3879.
- Zhang L, Chen X, Huang Y, Wu Z. 2019. The complete chloroplast genome of Primula helodoxa, a species endemic to China. Mitochondrial DNA B Resour. 5(1):194–195.
- Zhang C, Yuan X, Yang T, Yan H, Liu T. 2019. The complete chloroplast genome of Primula obconica (Primulaceae). Mitochondrial DNA B Resour. 4(2):2189–2190.
- Zhong L, Barrett SCH, Wang XJ, Wu ZK, Sun HY, Li DZ, Wang H, Zhou W. 2019. Phylogenomic analysis reveals multiple evolutionary origins of selfing from outcrossing in a lineage of heterostylous plants. New Phytol. 224(3):1290–1303.

