Abstract
Cuphea hookeriana Walp. is an ornamental plant belonging to the Lythraceae. In this study, we reported the complete chloroplast (cp) genome sequence here and analyzed the phylogenetic relationship among Lythraceae plants. The length of the cp genome was 158,999 bp, including a large single-copy (LSC, 89,311 bp) region and a small single-copy (SSC, 18,436 bp) region separated by a pair of inverted repeats (IRs, 25,626 bp). There were 72 unique protein-coding genes (PCGs), 30 transfer RNA (tRNA) genes, and four ribosomal RNA (rRNA) genes in the cp genome of C. hookeriana. A total of 223 simple sequence repeats (SSRs) and 34 long repeat sequences were identified. Phylogenetic analyses using maximum-likelihood (ML) revealed that C. hookeriana was close to C. hyssopifolia. In addition, the two Cuphea species were the sister group of Woodfordia fruticosa.
Introduction
Cuphea hookeriana Walp. 1897 is a member of the Lythraceae family () and originated in Mexico and distributed in tropical and south subtropical areas (Duss Citation1897). C. hookeriana is a small shrub with purple flowers that bloom all year. It is a popular ornamental plant that can be found in gardens, parks, and roadside green spaces and is now cultivated in Beijing, Tianjin, and Shaanxi, among other places in China. Besides, C. hookeriana is also identified as having economic value as an industrial oil plant, and the research about it is mainly focused on fatty acid synthases (Dehesh et al. Citation2001; Feng et al. Citation2018). Herein, in order to provide the basic genomic information for researching the evolution of Lythraceae, we described the entire chloroplast (cp) genome of C. hookeriana for the first time and carried out a phylogenetic analysis to resolve the relationship with close relatives.
Materials and methods
Fresh leaves samples of C. hookeriana were collected from the greenhouse of the College of Landscape and Architecture in Hangzhou (Hangzhou, China; 30°3′ N; 120°21′ E). The leaf of C. hookeriana was stored subsequently as a specimen at the Herbarium of Zhejiang A&F University under the accession code ZAFU20220106 (Cuihua Gu, [email protected]). The total DNA was extracted following the method of previous reports (Wang et al. Citation2021). Subsequently, the pair-end raw reads were produced based on the Illumina HiSeq platform at Beijing Genomics Institute (Shenzhen, China). After being purified by removing the adapter and low-quality reads, 3 Gb clean reads of C. hookeriana were used to perform the de novo assembly process. To obtain the complete cp genome of C. hookeriana, first, the reads of C. hookeriana were filtered by SeqKit (Shen et al. Citation2016). Subsequently, the resulting reads were assembled by SPAdes (Bankevich et al. Citation2012) using the cp genome of C. hyssopifolia (NC046574) as a reference. To know the accuracy of the assembly, we mapped the clean reads to assemble cp genome by Geneious Prime version 11.1.5 (Figure S1) (Deng et al. Citation2020). Subsequently, the assembled sequence was annotated and visualized by GeSeq (Tillich et al. Citation2017) after Bandage (Wick et al. Citation2015) made the necessary adjustments. In addition, the CPGVIEW (www.1kmpg.cn/cpgview/) (Liu et al. Citation2023) was applied to structures to visualize the intron-containing genes. The completed annotation was uploaded to GenBank and assigned the accession number OM949050. MISA (Beier et al. Citation2017) was used to search for simple sequence repeats (SSRs) of one to six nucleotide units (mono-, bi-, tri-, tetra-, penta-, and hexa-) across the cp genome. The minimal threshold of the unit size for searching was set as: 8, 5, 4, 3, 3, and 3. To find the long repeat sequences throughout the cp genome, REPuter (Kurtz et al. Citation2001) (https://bibiserv.cebitec.uni-bielefeld.de/reputer) was performed. The parameters are as follows: minimum size = 30, hamming distance = 3, and maximum computed repeats = 5000.
After aligning and trimming the cp genome by mafft v 7 (Katoh et al. Citation2019) and Gblocks 0.91b (Talavera and Castresana Citation2007), respectively, the IQtree v 1.6 (Nguyen et al. Citation2015) was used for generating a maximum-likelihood tree based on sequence from 14 Lythraceae species, three sequences from Onagraceae species were selected as an outgroup. The bootstrap was set to 1000 replicates, and the best-fit substitution model for the data (GTR + F + R2) was chosen by ModelFinder v 1.6.8 (Kalyaanamoorthy et al. Citation2017). The following sequences were used: NC037023 (Xue et al. Citation2017), MZ617461 (Fan et al. Citation2022), NC054309 (Lin et al. Citation2021), MN833212 (Wang et al. Citation2020), MK881637 (Gu et al. Citation2019), MK881626 (Gu et al. Citation2019), NC042891 (Gu et al. Citation2019), NC046574 (Ma et al. Citation2020), NC010358 (Gu et al. Citation2019), NC010361 (Chapman et al. Citation1999), NC029211 (Chapman et al. Citation1999), MK881638 (Gu et al. Citation2019), MG921615 (Gu et al. Citation2018), NC042897 (Xu et al. Citation2017), NC042896 (Xu et al. Citation2017), KF572028 (Wang et al. Citation2023), NC042890 (Wang et al. Citation2023), MH727532 (Jian and Ren Citation2019), and MK881631 (Gu et al. Citation2019).
Results
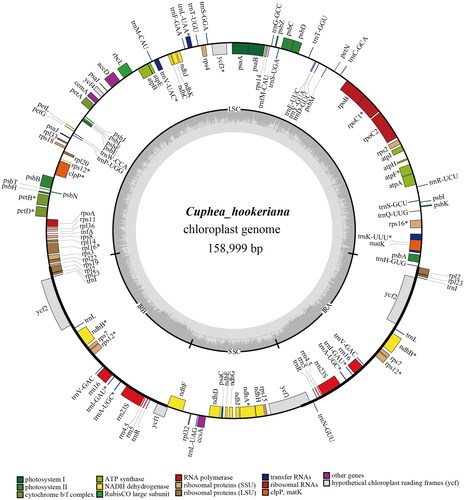
From the cp genome map of C. hookeriana (), the total length of the cp genome of C. hookeriana was 158,999 bp. A pair of inverted repeats (IRs, 25,626 bp) separated the large single-copy (LSC, 89,311 bp) region and small single-copy (SSC, 18,436 bp) region. LSC, SSC, and IRs contained the GC content of 36.97%, 31.05%, and 42.72%, respectively. A total of 106 unigenes were identified totally (44 of which were duplicated in the IRs) including 72 unique protein-coding genes (PCGs), 30 transfer RNA (tRNA) genes, and four ribosomal RNA (rRNA) genes. In addition, ycf 3, clpP, and ndhB had two introns (Figure S2). There 223 SSRs were found (Supple 1), including 196 mono-nucleotide, nine bi-nucleotide, seven tri-nucleotide, nine tetra-nucleotide, and four penta-nucleotide. With the threshold of e-value <1e–5, 34 long repeat sequences (Supple 2) were identified, consisting of 15 forward matches, 18 palindromic matches, and one reverse repeat.
Figure 2. Complete and regional genome map of C. hookeriana. The different colored legend below presents the different types of unigenes.
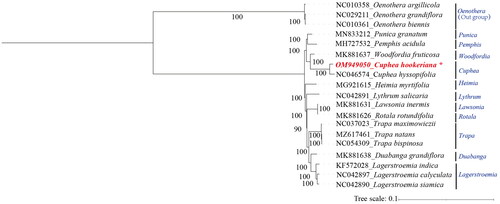
As shown in the phylogenetic tree (), the relationship of C. hookeriana in Lythraceae was presented. The C. hookeriana was mostly related to C. hyssopifolia with high bootstrap support, and as the sister group to Woodfordia fruticosa with C. hyssopifolia together.
Discussion and conclusions
C. hookeriana belongs to the Cuphea genus, and is a member of Lythraceae. Up to now, the research on phylogenetic analysis of the Lythraceae family has attracted much attention. The cp genome has been performed from many species of Lagerstroemia (Gu et al. Citation2016; Gu, Tembrock, Wu Citation2017; Gu, Tembrock, Zhang, et al. Citation2017), Heimia (Gu et al. Citation2018), Punica (Wang et al. Citation2020), and Trapa (Gu et al. Citation2019; Lin et al. Citation2021). In addition, the phylogenetic information of C. hyssopifolia has been published (Ma et al. Citation2020). However, the lack of cp genome sequence information from the tribe Cuphea makes it difficult to resolve the phylogenetic relationships of Cuphea species among the Lythraceae family. Herein, the cp genome of C. hookeriana was first sequenced and assembled to solve its relationship with other species in Lythraceae. As the result, C. hookeriana and C. hyssopifolia are most closely related, and they are the closest sister group to W. fruticosa.
Ethical approval
This research was carried out following the guidelines provided by the College of Landscape and Architecture, Zhejiang A&F University.
Author contributions
Sidan Hong, Yu Zhao, and Weili Shao were involved in the conception and design, Cuihua Gu, Yacheng Ye, and Guozhe Zhang participated in the analysis and drafting of the paper, as well as revised it critically for intellectual content; Mengxin Yu, Mingzhu Bai, and Qingqing Ma were involved in the interpretation of the data; and that all authors agree to be accountable for all aspects of the work.
Supplemental Material
Download JPEG Image (417.3 KB)Supplemental Material
Download MS Word (15.6 KB)Supplemental Material
Download MS Word (15.2 KB)Supplemental Material
Download JPEG Image (304.4 KB)Disclosure statement
No conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession OM949050. The numbers of BioProject, Biosample, and SRA number are PRJNA814813, SAMN26567440, and SRR18298639, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Beier S, Thiel T, Münch T, Scholz U, Mascher M. 2017. MISA-web: a web server for microsatellite prediction. Bioinformatics. 33(16):2583–2585.
- Chapman MJ, Mulcahy DL, Stein DB. 1999. Identification of plastid genotypes in Oenothera subsect. Munzia by restriction fragment length polymorphisms (RFLPs). Theor Appl Genet. 98(1):47–53.
- Dehesh K, Tai H, Edwards P, Byrne J, Jaworski JG. 2001. Overexpression of 3-ketoacyl-acyl-carrier protein synthase IIIs in plants reduces the rate of lipid synthesis. Plant Physiol. 125(2):1103–1114.
- Deng X, Gu W, Federman S, Du Plessis L, Pybus OG, Faria NR, Wang C, Yu G, Bushnell B, Pan C-Y, et al. 2020. Genomic surveillance reveals multiple introductions of SARS-CoV-2 into Northern California. Science. 369(6503):582–587.
- Duss R-PA. 1897. Flore phanérogamique des Antilles françaises.
- Fan X, Wang W, Wagutu GK, Li W, Li X, Chen Y. 2022. Fifteen complete chloroplast genomes of Trapa species (Trapaceae): insight into genome structure, comparative analysis and phylogenetic relationships. BMC Plant Biol. 22(1):230.
- Feng Y, Zhang Y, Wang Y, Liu J, Liu Y, Cao X, Xue S. 2018. Tuning of acyl-ACP thioesterase activity directed for tailored fatty acid synthesis. Appl Microbiol Biotechnol. 102(7):3173–3182.
- Gu C, Dong B, Xu L, Tembrock LR, Zheng S, Wu Z. 2018. The complete chloroplast genome of Heimia myrtifolia and comparative analysis within Myrtales. Molecules. 23(4):846.
- Gu C, Ma L, Wu Z, Chen K, Wang Y. 2019. Comparative analyses of chloroplast genomes from 22 Lythraceae species: inferences for phylogenetic relationships and genome evolution within Myrtales. BMC Plant Biol. 19(1):281.
- Gu C, Tembrock LR, Johnson NG, Simmons MP, Wu Z. 2016. The complete plastid genome of Lagerstroemia fauriei and loss of rpl2 intron from Lagerstroemia (Lythraceae). PLOS One. 11(3):e0150752.
- Gu C, Tembrock LR, Wu Z. 2017. The complete chloroplast genome of Lagerstroemia intermedia (Lythraceae), a threatened species endemic to southwestern Yunnan province, China. Conserv Genet Resour. 9(3):357–360.
- Gu C, Tembrock LR, Zhang D, Wu Z. 2017. Characterize the complete chloroplast genome of Lagerstroemia floribunda (Lythraceae), a narrow endemic crape myrtle native to Southeast Asia. Conserv Genet Resour. 9(1):91–94.
- Jian S, Ren H. 2019. The complete chloroplast genome sequence of Pemphis acidula (Lythraceae). Mitochondrial DNA B. 4(1):912–913.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. 2001. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29(22):4633–4642.
- Lin L, Wang J, Zhao YX, Ma L, Gu CH, Wu ZQ. 2021. Chloroplast genome of Trapa bispinosa Roxb. (Trapa, Lythraceae). Mitochondrial DNA B Resour. 6(2):333–334.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704.
- Ma L, He Y-Z, Yu J-Y, Gu C-H, Liao X-Z, Wu Z-Q. 2020. The complete chloroplast genome sequence of Cuphea hyssopifolia. Mitochondrial DNA B. 5(2):1415–1416.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Shen W, Le S, Li Y, Hu F. 2016. SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLOS One. 11(10):e0163962.
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wang J, He W, Liao X, Ma J, Gao W, Wang H, Wu D, Tembrock LR, Wu Z, Gu C. 2023. Phylogeny, molecular evolution, and dating of divergences in Lagerstroemia using plastome sequences. Hortic Plant J. 9(2):345–355.
- Wang J, Wu Z, Ma L, Gu C. 2021. Characterization of the chloroplast genome of Lagerstroemia villosa Wall. ex Kurz. (Lagerstroemia, Lythraceae). Mitochondrial DNA B Resour. 6(1):19–20.
- Wang J, Wu ZQ, Ma L, Gu CH. 2020. Complete chloroplast genome sequence of Punica granatum 'Nana’ (Lythraceae) and phylogenetic analysis. Mitochondrial DNA B Resour. 5(3):2070–2071.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352.
- Xu C, Dong W, Li W, Lu Y, Xie X, Jin X, Shi J, He K, Suo Z. 2017. Comparative analysis of six Lagerstroemia complete chloroplast genomes. Front Plant Sci. 8:15.
- Xue Z-Q, Xue J-H, Victorovna KM, Ma K-P. 2017. The complete chloroplast DNA sequence of Trapa maximowiczii Korsh. (Trapaceae), and comparative analysis with other Myrtales species. Aquat Bot. 143:54–62.