Abstract
The Korean endemic Eranthis byunsanensis B.Y. Sun, Citation1993 (Ranunculaceae) is a rare plant distributed in the southwestern part of the Korean Peninsula. The complete chloroplast (cp) genome of E. byunsanensis was sequenced by next-generation sequencing (NGS) using an Illumina HiSeq X platform. The cp genome of E. byunsanensis is 160,324 bp in length with 37.9% GC content. It showed a typical quadripartite structure consisting of a pair of inverted repeats (IRs; 28,356 bp), a large single-copy region (LSC; 87,671 bp), and a small single-copy region (SSC; 15,941 bp). The cp genome comprises 130 genes including 85 protein-coding genes (PCGs), 37 tRNA genes, and eight rRNA genes. The molecular phylogenetic analysis indicates that E. byunsanensis is closely related to Eranthis stellata, both of which belong to the genus Eranthis.
Introduction
The genus Eranthis Salisb., an early flowering perennial plant belonging to Ranunculaceae Juss. Tribe Cimicifugeae Torr. & A. Grey, has been a subject of interest in phylogenetic research because it is often considered the most primitive of herbaceous flowering plant (Tamura Citation1995). This genus comprises approximately 13 species that are mainly distributed in East Asia and Europe (Tamura Citation1995). Of these, three Eranthis species including E. stellata Maxim., 1859, E. byunsanensis B.Y.Sun, Citation1993, and E. pungdoensis B.U.Oh, 2009 inhabit limited areas in the Korean peninsula. In particular, E. byunsanensis and E. pungdoensis were recently described as new species (Sun et al. Citation1993; Oh and Ji Citation2009). However, the phylogenetic position of these species has been controversial due to their high similarities in taxonomic characteristics. The shapes of petals, leaves, and bracts showed only minor differences between these species (Oh and Oh Citation2019).
Chloroplast (cp) genome sequences are precious resources for phylogenetic study on closely related taxa (Bi et al. Citation2018). To date, the complete cp genome sequences of E. stellata have been only reported among 13 Eranthis species (Zhai et al. Citation2019). Here, we analyzed the complete cp genome sequence of E. byunsanensis and evaluated its phylogenetic position within the family Ranunculaceae. Our results may provide valuable resources for future genetic and evolutionary studies on Ranunculaceae.
Materials and methods
Plant sampling
The plant material of E. byunsanensis was collected from the mountain area of Wanju, Jeollabuk-do, South Korea (127°18′15.50″E, 36°07′17.20″N) on 22 March 2022 and photographed with a digital camera to record the natural habitat (). Each specimen was morphologically identified by Joon Moh Park (https://forest.jb.go.kr/, [email protected]). The specimen voucher with number JFERI0020-1 was deposited in the Jeollabuk-do Forest Environment Research Institute.
Figure 1. Eranthis byunsanensis of Wanju, Republic of Korea. (A) Typical radical leaves with blades divided into several palmate segments. (B) White, compound floral structure with broad overlapping sepals and funnel-shaped petals. Scale bars represent 1 cm. Photographs were captured by Joon Moh Park using a digital camera.

Sequencing, assembling, and annotating the chloroplast genome
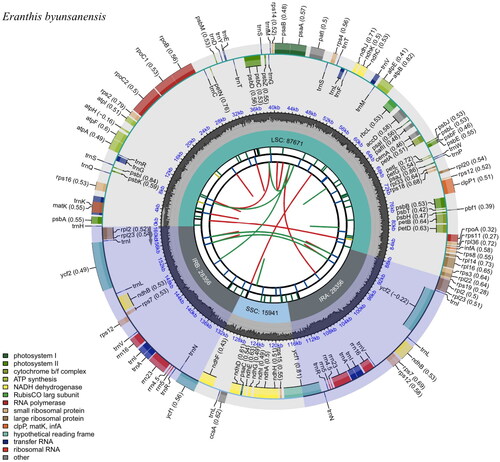
Total genomic DNA was isolated from the radical leaves of E. byunsanensis and deposited in the Jeollabuk-do Forest Environment Research Institute (voucher number JFERI-DNA0020-1; contact person, Joon Moh Park). The next-generation sequencing (NGS) sequencing library was constructed using TruSeq Nano DNA Kit (Illumina, San Diego, CA) and sequenced by paired-end sequencing using Illumina HiSeq X platform (Macrogen Inc., Seoul, Republic of Korea). Trimmomatic was used to eliminate adapter sequences and low-quality reads. De novo assembly was performed using NOVOPlasty v.4.3.1 (Dierckxsens et al. Citation2017). Annotation of cp genome was conducted by GeSeq v.1.59 (https://chlorobox.mpimp-golm.mpg.de/geseq.html). The cis- and trans-splicing genes were identified using CPGView software (Liu et al. Citation2023). A circular map of the complete cp genome was constructed by CPGView software.
Phylogenetic analysis
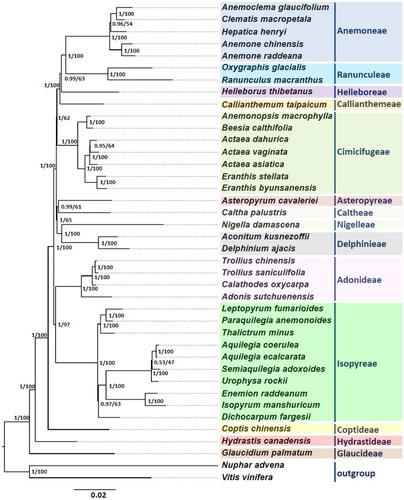
The phylogenetic position of E. byunsanensis was investigated using 38 species of Ranunculaceae and two outgroup species. The nucleotide sequences of 77 shared non-redundant protein-coding genes (PCGs) were extracted from the complete cp genome sequences deposited in GenBank. The PCGs were concatenated into a dataset using Geneious prime 2023 (Biomatters Ltd, Auckland, New Zealand). Multiple sequence alignment was performed using MAFFT (Katoh and Standley Citation2013), and gaps and poorly aligned sequences were eliminated by TrimmAl. The resulting 75,566 bp of aligned sequences were used to generate a phylogenetic tree using two methods. The maximum-likelihood (ML) method was performed by RAxML (Stamatakis Citation2014) with 1000 replicates using the GTR + G model. Bayesian’s inference (BI) method was performed using MrBayes (Ronquist et al. Citation2012) with the GTR + I + G model. Each analysis implemented four MCMC chains with 5,000,000 generations and sampled every 1000 iterations with the first 10% discarded.
Results
Chloroplast genome features
The cp genome of E. byunsanensis was sequenced by paired-end sequencing. The 28,939,092 filtered sequencing reads were assembled into single contigs with an average read coverage of 1187 (Supplementary Figure S1). The assembly result was evaluated by depth of coverage using Bowtie2 and verified by comparing the synteny and sequence homology with the reference cp sequence (Eranthis stellate, MK569487) by NOVOPlasty v.4.3.1 (Supplementary Figure S1). PCR-based Sanger sequencing was performed to confirm the complete circular cp genome sequences (Supplementary Figure S2). A schematic representation of the plastome organization of E. byunsanensis is represented in . The complete cp genome of E. byunsanensis (GenBank accession number ON564441) is 160,324 bp in length with an overall GC content of 37.9%. It showed a typical quadripartite structure comprising a pair of inverted repeats (IRA and IRB; 28,356 bp), a large single-copy region (LSC; 87,671 bp), and a small single-copy region (SSC; 15,941 bp). A total of 130 genes were identified, including 85 PCGs, 37 tRNA genes, and eight rRNA genes. Among the PCGs, 13 contain one or two introns (Supplementary Figure S3). The rps12 is a trans-splicing gene with 5′-end and 3′-end exons located in the LSC and IRA regions, respectively.
Figure 2. Chloroplast genome map of E. byunsanensis. A circular and complete cp genome map was generated by CPGview. Large single-copy, small single-copy, and inverted repeat are represented as LSC, SSC, and IR (IRA and IRB) on the fourth track, respectively. GC content is represented on the fifth track in dark gray. The genes are shown on the sixth track. Genes located on the inner and outer of circle are transcribed clockwise and anticlockwise, respectively. The functional classification of the genes is presented in the bottom left corner.
Phylogenetic analysis
Phylogenetic position of E. byunsanensis was analyzed using the 77 conserved PCGs in the complete cp genomes of 38 Ranunculaceae species including E. byunsanensis and two outgroups (). Two phylogenetic trees were constructed using BI and ML methods. BI and ML trees showed identical topologies with 14 strongly supported clades, each of which corresponds to the defined tribe in previous studies (Zhai et al. Citation2019). In addition, the phylogenetic tree indicates that E. byunsanensis is most closely related to E. stellata with strong support (posterior probability = 1/bootstrap = 100).
Figure 3. Phylogenetic relationships among 38 Ranunculaceae species based on 77 shared PCGs of the complete cp genome. The sequences used for tree construction are as follows: Anemoclema glaucifolium (MK569471; Zhai et al. Citation2019), Anemone chinensis (MK569491; Zhai et al. Citation2019), Anemone raddeana (MK569472; Zhai et al. Citation2019), Clematis macropetala (MK569482; Zhai et al. Citation2019), Hepatica henryi (MK569494; Zhai et al. Citation2019), Oxygraphis glacialis (MK569489; Zhai et al. Citation2019), Ranunculus macranthus (NC_008796; Raubeson et al. Citation2007), Helleborus thibetanus (MK569493; Zhai et al. Citation2019), Callianthemum taipaicum (MK569479; Zhai et al. Citation2019), Actaea asiatica (MK569469; Zhai et al. Citation2019), Actaea dahurica (MK569481; Zhai et al. Citation2019), Actaea vaginata (MK569499; Zhai et al. Citation2019), Anemonopsis macrophylla (MK569473; Zhai et al. Citation2019), Beesia calthifolia (MK569477), Eranthis stellate (MK569487; Zhai et al. Citation2019), Eranthis byunsanensis (ON564441; this study), Caltha palustris (MK569480; Zhai et al. Citation2019), Asteropyrum cavaleriei (MK569476; Zhai et al. Citation2019), Nigella damascene (MK569488; Zhai et al. Citation2019), Aconitum kusnezoffii (MK569468; Zhai et al. Citation2019), Delphinium ajacis (MK569484; Zhai et al. Citation2019), Adonis sutchuenensis (MK569470; Zhai et al. Citation2019), Calathodes oxycarpa (MK569478; Zhai et al. Citation2019), Trollius saniculifolia (NC_012615; Kim et al. Citation2009), Trollius chinensis (MK569501; Zhai et al. Citation2019), Aquilegia coerulea (MK569474; Zhai et al. Citation2019), Aquilegia ecalcarata (MK569475; Zhai et al. Citation2019), Dichocarpum fargesii (MK569485; Zhai et al. Citation2019), Enemion raddeanum (MK569486; Zhai et al. Citation2019), Isopyrum manshuricum (MK569496; Zhai et al. Citation2019), Leptopyrum fumarioides (MK569497; Zhai et al. Citation2019), Paraquilegia anemonoides (MK569490; Zhai et al. Citation2019), Semiaquilegia adoxoides (MK569498; Zhai et al. Citation2019), Thalictrum minus (MK569500; Zhai et al. Citation2019), Urophysa rockii (MK569502; Zhai et al. Citation2019), Coptis chinensis (MK569483; Zhai et al. Citation2019), Hydrastis Canadensis (MK569495; Zhai et al. Citation2019), Glaucidium palmatum (MK569492; Zhai et al. Citation2019). Nuphar advena (NC_008788; Raubeson et al. Citation2007), and Vitis vinifera (NC_007957; Jansen et al. Citation2006) were used as outgroups. The phylogenetic tree was constructed using BI and ML methods, and the trees showed identical topologies. The numbers on the nodes indicate the BI posterior probability and ML bootstrap value (%), respectively. The scale bar represents the number of substitutions per site.
Discussion and conclusions
The family Ranunculaceae has been the focus of systematic studies as a model due to its unstable position in flowering plants and diverse morphological characteristics (Tamura Citation1995). Recently, this family has received more attention as a model system for addressing crucial evolutionary questions regarding either primitive or derived morphological features that cannot be answered by other model plants (Kramer Citation2009). Numerous studies have attempted to determine the divergence times among the major lineages of Ranunculaceae, but no consensus has been achieved (Wang et al. Citation2016). In particular, the phylogenetic position of the genus Eranthis has been controversial due to their high similarities in taxonomic characteristics. Recently, the cp genomes of 35 Ranunculaceae species representing 31 genera of the 14 tribes were phylogenetically analyzed, and inter-tribal relationships of the family Ranunculaceae were much clarified (Zhai et al. Citation2019). In the present study, the complete cp genome sequences of E. byunsanensis were determined and utilized to analyze its phylogenetic position within 38 Ranunculaceae species. The BI and ML phylogenetic trees strongly indicate that E. byunsanensis is most closely related to E. stellata, both of which belong to the genus Eranthis. Our data provide valuable information for further genetic and evolutionary studies on Ranunculaceae.
Author contributions
J.M.P., A.O., and J.K. carried out sample collection, experiments, data analysis, and data curation. Preparation of manuscript and project administration were performed by J.K. All authors have read and agreed to the published version of the manuscript.
Ethical approval
The species used in this study does not need ethical approval or permissions to collect the sample. All procedures for the sampling and experiments in this article were conducted in compliance with the regulations of the Jeollabuk-do Forest Environment Research Institute.
Supplemental Material
Download MS Word (843.5 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data supporting the findings of this study are openly available as accession no. ON564441 at GenBank in NCBI (https://www.ncbi.nlm.nih.gov). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA835661, SRR19213060, and SAMN28106645, respectively.
Additional information
Funding
References
- Bi Y, Zhang MF, Xue J, Dong R, Du YP, Zhang XH. 2018. Chloroplast genomic resources for phylogeny and DNA barcoding: a case study on Fritillaria. Sci Rep. 8(1):1184.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Jansen RK, Kaittanis C, Saski C, Lee SB, Tomkins J, Alverson AJ, Daniell H. 2006. Phylogenetic analyses of Vitis (Vitaceae) based on complete chloroplast genome sequences: effects of taxon sampling and phylogenetic methods on resolving relationships among rosids. BMC Evol Biol. 6:32.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kramer EM. 2009. Aquilegia: a new model for plant development, ecology, and evolution. Annu Rev Plant Biol. 60:261–277.
- Kim YK, Park CW, Kim KJ. 2009. Complete chloroplast DNA sequence from a Korean endemic genus, Megaleranthis saniculifolia, and its evolutionary implications. Mol Cells. 27(3):365–381.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704.
- Oh BU, Ji SJ. 2009. Eranthis pungdoensis B.U. Oh: a new species of Eranthis (Ranunculaceae) from Korea. Korean J Plant Taxon. 39(2):86–88.
- Oh A, Oh BU. 2019. The speciation history of northern- and southern-sourced Eranthis (Ranunculaceae) species on the Korean peninsula and surrounding areas. Ecol Evol. 9(5):2907–2919.
- Raubeson LA, Peery R, Chumley TW, Dziubek C, Fourcade HM, Boore JL, Jansen RK. 2007. Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genomics. 8:174.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Sun BY, Kim CH, Kim TJ. 1993. A new species of Eranthis (Ranunculaceae) from Korea: E. byunsanensis. Korean J Plant Taxon. 23(1):21–26.
- Tamura M. 1995. Phylogeny and classification of the Ranunculaceae. Plant Syst Evol Suppl. 9:201–206.
- Wang W, Lin L, Xiang XG, Ortiz RDC, Liu Y, Xiang KL, Yu SX, Xing YW, Chen ZD. 2016. The rise of angiosperm-dominated herbaceous floras: insights from Ranunculaceae. Sci Rep. 6:27259.
- Zhai W, Duan X, Zhang R, Guo C, Li L, Xu G, Shan H, Kong H, Ren Y. 2019. Chloroplast genomic data provide new and robust insights into the phylogeny and evolution of the Ranunculaceae. Mol Phylogenet Evol. 135:12–21.
