Abstract
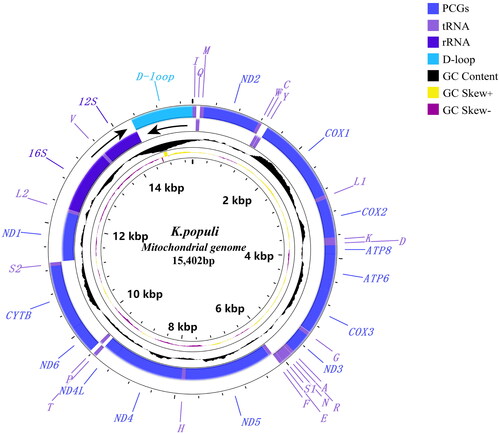
In this study, we sequenced and reported the complete mitochondrial genomes of Kusala populi for the first time. The complete mitochondrial genome was registered in GenBank with accession number NC_064377 as the first complete mitogenome of the genus Kusala. The circular mitochondrial genome length is 15,402 bp, with nucleotide composition A (41.8%), C (11.4%), G (9.2%), T (37.6%), A + T (79.4%), and C + G (20.6%), comprising 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes and a D-loop region. All protein-coding genes were encoded by the H-strand, except for 4 genes (nad5, nad4, nad4L, nad1). 8 tRNA genes (tRNA-Gln, tRNA-Cys, tRNA-Tyr, tRNA-Phe, tRNA-His, tRNA-Pro, tRNA-Leu, tRNA-Val) and 2 rRNA genes (16S, 12S) were encoded in the L-strand. Phylogenetic analysis indicated that the newly sequenced species had a close relationship with Mitjaevia, another widespread Old-World genus of Erythroneurini.
1. Introduction
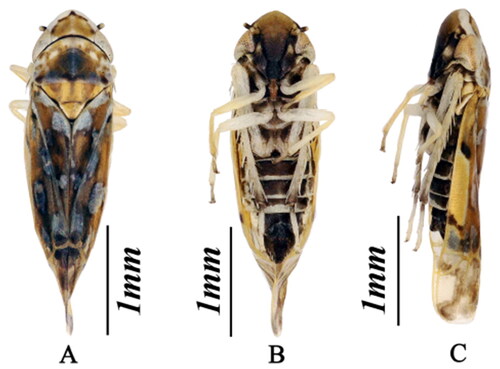
Leafhoppers, phytophagous insects, are important pests of agriculture, forestry, fruit trees, and other economically important plants (Morris Citation1971; Guo Citation2011; Roddee et al. Citation2018). Cicadellidae is the largest family in Hemiptera, and is distributed all over the worldwide (Zhang et al. Citation2021). Typhlocybinae is one of the important forestry pests of Cicadellidae. There are eleven species of Oriental genus Kusala widely distributed in the world, among them, four are in China, including Kusala colibria, Kusala datianensis, Kusala maculata and Kusala populi (Song and Li Citation2014). K. populi (Song et al. Citation2011) were found in karst areas in Guizhou and Chongqing. However, genetic information on this species has not yet been studied. Thus, we sequenced the mitogenome of K. populi to determine the phylogenetic position of the species and the genus Kusala. As a first report on the mitogenome of genus Kusala, this result will be useful in the future for species identification, phylogenetic studies and biogeographic studies within Typhlocybinae.
2. Materials and methods
2.1. Sample collection
In mid-July 2019, K. populi adult specimens (twenty female individuals, ten male individuals, ) were collected using the sweeping net method from roadside weeds in Huaguan Road, Huaxi District, Guiyang City (26°24′44.90″N, 106°41′33.70″°E, elevation 1171 m). The specimens were morphologically identified, stored in absolute ethanol, and placed in a freezer at −20 °C. The sample and the total DNA were deposited at School of Karst Science, Guizhou Normal University (Voucher number: GZNU-ELS-20190715, http://gznu.edu.cn, contact person: Weiwen Tan; email: [email protected]).
2.2. Methods
The leafhoppers total DNA was extracted from the entire body of multiple males of the same species (seven individual males of K. populi) without the abdomen and wings. Genomic DNA was extracted by using a tissue rapid Extraction Kit (VWI) according to the manufacturer’s instructions. Whole mitochondrial genome sequences were determined at Berry Genomics (Beijing, China) using an Illumina Novaseq 6000 platform (Illumina, Alameda, CA, USA) by 150 bp paired-end reads and 5.87 G of raw data were obtained. Mitochondrial genome assembly was performed using GetOrganelle 1.7.5 (parameters: -R 10 -k 21,45,65,85,105 -F animal_mt) (Jin et al. Citation2020), and the assembly results were blastn (version: BLAST 2.2.30+; parameters: – evalue 1e-5) with the proximal reference mitogenome (accession number NC_047465), and the candidate sequence assembly results were determined based on the comparison. Gene annotation of the assembled mitogenome was performed using MITOS 2 (Alexander et al. Citation2019). The annotated mitogenome was deposited in GenBank with the accession number NC_064377. In order to understand the phylogenetic status of K. populi, we downloaded 20 closely related mitogenomes from the NCBI database, the Homalodisca coagulate and Cicadella viridis mitogenomes were selected as the outgroup (GenBank no. AY875213 and no. MK335936). Maximum likelihood (ML) method in IQ-TREE 2.0 (Minh et al. Citation2020; 2000 ultrafast bootstrap) based on the best-fit model GTR + I + R and Bayesian inference (BI) method in MrBayes 3.2 (Ronquist et al. Citation2012) were adopted to construct phylogenetic tree using the complete mitochondrial genes.
3. Results
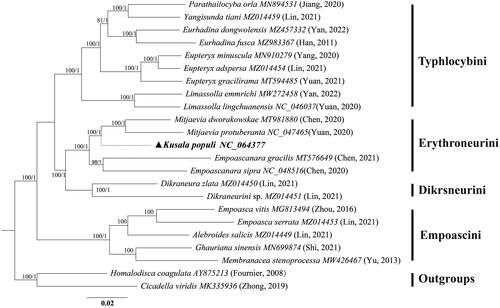
The mitochondrial genome of K. populi was 15,402 bp in length, containing 13 protein-coding genes (PCGs), 22 tRNAs, 2 rRNA genes, and a D-loop region (). The nucleotide composition of the genome is A (41.8%), C (11.4%), G (9.2%), T (37.6%), A + T (79.4%), and C + G (20.6%). There were 14 genes encoded in the L-strand, and the remaining genes were encoded in the H-strand. The largest gene was nad5 and the smallest was atp8 in PCGs. The AT content in PCGs was slightly lower than that in the whole genome. All PCGs start from the ATN (ATA/ATT/ATG) codon except atp8 where it was TTG and end with the complete stop codon TAA or TAG, TAA was the most frequently used stop codon. The cox2, nad1, and nad4 have an incomplete stop codon T–, and the tRNA structure of K. populi is similar to that of other Hemiptera (Yuan et al. Citation2021). The 21 tRNAs can be folded into a typical cloverleaf secondary structure, except trnS1, which lacks the dihydrouridine (DHU) stem and forms a simple loop. Phylogenetic analysis indicated that Kusala belongs to the tribe Erythroneurini, a sister clade to Mitjaevia, i.e. ((Kusala + Mitjaevia) + Empoascanara) () (; UBP/BPP= 100/1.00).
4. Discussion and conclusion
The mitochondrial genome of K. populi was 15,402 bp in length. It contained 13 protein-coding genes (PCGs), 22 tRNAs, 2 rRNA genes, and a D-loop region (). The analysis showed that the genomic structure of K. populi is similar to previously studied (Chen et al. Citation2021). Reconstructed phylogenetic trees based on 21 subfamily Typhlocybinae species and two outgroups yielded a consistent topology with similar phylogenetic relationships between clades as in previous studies, i.e. (Empoascini + (Typhlocybini + (Erythroneurini + Dikrsneurini))). Our study reveals for the first time the phylogenetic position of Kusala within the tribe Erythroneurini and suggests a closer genetic relationship with Mitjaevia, which is consistent with the results of Song et al. (Citation2011). Our phylogenetic tree also shows the robustness of the mitochondrial genome in resolving phylogenetic relationships at the tribe and genus level due to the high level of support obtained for these nodes (; UBP/BPP = 100/1.00). The resultwill contribute to future phylogenetic studies, population genetics, and biogeography within the subfamily Typhlocybinae.
Ethical approval
The research of the article does not need Ethical approval. No specific permissions were required for any of the collection localities.
Authors’ contributions
Tianyi Pu: conceived and designed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Ni Zhang: analyzed the data, prepared figures and/or tables, and approved the final draft.
Yu Zhang: analyzed the data, reviewed drafts of the paper, prepared figures and/or tables, and approved the final draft.
Yuehua Song: conceived and designed the experiments, species identification and authored or reviewed drafts of the paper, and approved the final draft.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in the GenBank of NCBI athttps://www.ncbi.nlm.nih.gov under the accession NC_064377. The associated BioProject, SRA and Bio-sample numbers are PRJNA842561, SRR19414868, and SAMN28677931, respectively.
Additional information
Funding
References
- Alexander D, Frank J, Marwa AA, Bernhart SH, Franziska R, Stadler PF, Middendorf M, Bernt M. 2019. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 47(20):10543–10552.
- Chen XX, Li C, Song YH. 2021. The complete mitochondrial genomes of two erythroneurine leafhoppers (Hemiptera, Cicadellidae, Typhlocybinae, Erythroneurini) with assessment of the phylogenetic status and relationships of tribes of Typhlocybinae. Zookeys. 1037:137–159.
- Guo H. 2011. Major tea pests-advances in research on pseudo-eye leafhopper. Jiangsu Agric Sci. 1:132–134.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):1–31.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Morris MG. 1971. Differences between the invertebrate faunas of grazed and ungrazed chalk grassland. IV. Abundance and diversity of Homoptera-Auchenorrhyncha. J Appl Entomol. 8(1):37–52.
- Roddee J, Kobori Y, Hanboonsong Y. 2018. Multiplication and distribution of sugarcane white leaf phytoplasma transmitted by the leafhopper, Matsumuratettix hiroglyphicus (Matsumura) (Hemiptera: Cicadellidae), in infected sugarcane. Sugar Tech. 20(4):445–453.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Song YH, Li ZZ, Xiong KN. 2011. New species and new records of Erythroneurini from China (Hemiptera: Cicadellidae: Typhlocybinae). Zootaxa. 2774(1):48–56.
- Song YH, Li ZZ. 2014. Erythroneurini and Zyginellini from China (Hemiptera: Cicadellidae: Typhlocybinae). Guiyang: Guizhou Science and Technology Publishing House.
- Zhou X, Dietrich CH, Huang M. 2020. Characterization of the complete mitochondrial genomes of two species with preliminary investigation on phylogenetic status of Zyginellini (Hemiptera: Cicadellidae: Typhlocybinae). Insects. 11(10):684.
- Yuan ZW, Xiong KN, Zhang N, Li C, Song Y. 2021. Characterization of the morphology and complete mitochondrial genomes of Eupteryx minusula and Eupteryx gracilirama (Hemiptera: Cicadellidae: Typhlocybinae) from Karst area. Southwest China. 9:e12501.
- Zhang N, Song QF, Song YH. 2021. A new leafhopper genus of Erythroneurini (Hemiptera, Cicadellidae, Typhlocybinae) from karst area in southwestern China. Rev Bras Entomol. 65(4):e20210041.