Abstract
Syringa oblata var. alba is a shrub or a small tree from China with high ornamental, medicinal, and edible value. Here, we present its first complete chloroplast genome. The entire circular genome is 155,648 bp in length, with large single-copy (LSC) length of 86,247, small single-copy (SSC) length of 17,937, inverted repeat (IR) length of 25,732, and GC content of 37.9%. One hundred and thirty-two genes, including 88 protein-coding, 36 tRNA, and eight rRNA genes were predicted. A phylogenetic tree of 25 plant species was constructed based on the maximum-likelihood method, indicating that S. oblata var. alba, S. vulgaris, and S. oblata form a sister group. This study will provide valuable basic information for phylogeny, species identification, and varieties breeding of this species.
1. Introduction
Syringa oblata Lindl. var. alba Hort. ex Rehd. 1763, a shrub or a small tree in Syringa genus, has strong adaptability to cold, drought, barren environment, and resistance to diseases and insect pests. It is distributed in the north of the Yangtze River Basin in China, with flowering period from April to May and fruit period from June to October, according to the record of Flora Reipublicae Popularis Sinicae (FRPS). S. oblata var. alba is generally used for garden cultivation and ornamental for its lush branches and leaves, elegant flowers and fragrant smell. Moreover, the flowers of many Syringa plants are the materials of spices and clove oil which can be used in cosmetics, drugs, and food (Cheng et al. Citation2021). Taxonomically, Syringa is closely related to Ligustrum. Sect. Ligustrina of Syringa possesses similar floral and seed characteristics to some species of Ligustrum. This finding is consistent with the report of nuclear ribosomal DNA ITS and ETS region sequencing to test Syringa and Ligustrum relationship (Li et al. Citation2002). In addition, a study of S. oblata genome pointed out that segment and tandem duplications could contribute to the odor formation in the woody aromatic species (Wang et al. Citation2022). In this study, the complete chloroplast (cp) genome of S. oblata var. alba was assembled and characterized for the first time, which would enrich the genomic resources of Oleaceae and contribute to the identification and application of Syringa.
2. Materials and methods
S. oblata var. alba () was sampled from Dalian, Liaoning, China (E 121°35′39″, N 38°58′13″) and identified by professor Ting-guo Kang. The voucher specimen and genomic DNA were deposited at the herbarium of Liaoning University of Traditional Chinese Medicine (Liang Xu [email protected], plant number: 10162210515096LY). All operations are carried out in accordance with guidelines in Specification on Good Agriculture and Collection Practices for Medicinal Plants (GACP; Number: T/CCCMHPIE 2.1-2018). The extraction of total genomic DNA from fresh leaves was achieved by Magbead Plant DNA Kit (CWBIO, Taizhou, China) and sequenced on Illumina Novaseq 6000 platform. Data were processed and assembled by NGS QC toolkit (Patel and Jain Citation2012) and SPAdes v3.14.1 (Bankevich et al. Citation2012), respectively. The protein-coding sequences of cp genome were compared with NR (RefSeq non-redundant proteins) databases for the gene prediction and annotation.
3. Results and discussion
The cp genome of S. oblata var. alba was 155,648 bp in length with a GC content of 37.9%. One hundred and thirty-two genes were predicted, including 88 protein-coding, 36 tRNA, and eight rRNA genes. The genome had a large single-copy (LSC) region with a length of 86,247 bp, a small single-copy (SSC) region of 17,937 bp, and a pair of inverted repeats (IRs) regions of 25,732 bp. In addition, trnK-UUU, rps16, trnG-GCC, atpF, rpoC1, trnL-UAA, petB, petD, rpl16, rpl2, ndhB, trnI-GAU, trnA-UGC, and ndhA contain a single intron, clpP and ycf3 genes contain two introns, and rps12 gene has trans splicing. As the evidence for correct assembly of the genome, coverage depth figure was provided in Supplemental Figure S1. The maps of annotated cp genome, cis-splicing genes, and trans-splicing genes of S. oblata var. alba were processed by CPGview (Liu et al. Citation2023) and shown in , , respectively.
Figure 2. The annotated chloroplast genome map of S. oblata var. alba. From the center outward, the forward and reverse repeats were shown in the first track and connected with green and red arcs, respectively. The tandem repeats and microsatellite sequences were shown in the second and third tracks with different colors. Genes are colored in the outer track by their functional classification listed in the bottom left.
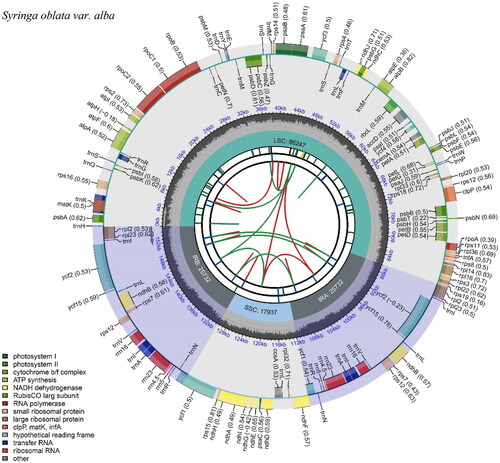
Figure 3. Maximum-likelihood (ML) phylogenetic tree based on the complete chloroplast genome of S. oblata var. alba and 24 other species. The following sequences were used.
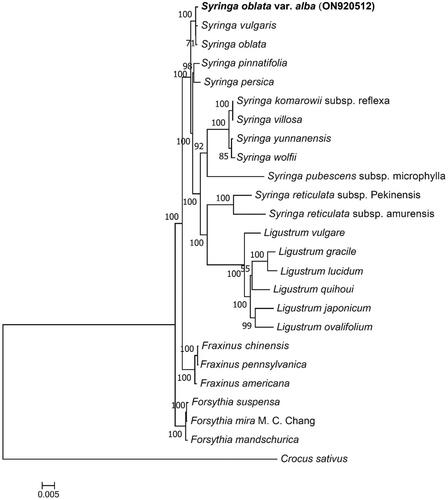
Maximum-likelihood (ML) phylogenetic tree was conducted on 25 species of plant. The ML analysis was performed by IQ-TREE 1.6.12 software (http://www.iqtree.org/) with 1000 bootstrap replicates (Nguyen et al. Citation2015). According to the Bayesian information criterion (BIC), the best evolutionary model was chosen as JTT + F + R3. Based on this phylogenetic tree, all selected species in Ligustrum are located in a branch within Syringa branches, indicating the close relationship between Ligustrum and Syringa. S. oblata var. alba, S. vulgaris, and S. oblata are sister groups, and the relationship between the latter two is consistent with a previous study (Zhao et al. Citation2020). Crocus sativus, as an outgroup, is far from the other species ().
Syringa vulgaris MG255768, S. oblata NC057990, S. pinnatifolia NCO41119 (Zhang et al. Citation2019), S. persica NC042280 (Olofsson et al. Citation2019), S. komarowii subsp. reflexa MT648823, S. villosa NC061378, S. yunnanensis NC042468 (Olofsson et al. Citation2019), S. wolfii NC049090 (Liu et al. Citation2020), S. pubescens subsp. microphylla MT872641, S. reticulata subsp. pekinensis MN901632 (Wang et al. Citation2020), S. reticulata subsp. amurensis MT872640, Ligustrum vulgare NC042274 (Olofsson et al. Citation2019), L. gracile NC042425 (Olofsson et al. Citation2019), L. lucidum NC056243, L. quihoui NC057246 (Wang et al. Citation2019), L. japonicum NCO42454 (Olofsson et al. Citation2019), L. ovalifolium NC056242, Fraxinus chinensis MW599993, F. pennsylvanica NCO43874 (Yi et al. Citation2019), F. americana NCO42449 (Olofsson et al. Citation2019), Forsythia suspensa NC036367 (Wang et al. Citation2017), F. mira M. C. Chang NC046065 (Gao et al. Citation2019), F. mandschurica NC048504 (Olofsson et al. Citation2019), and Crocus sativus NC041460 (Nemati et al. Citation2019).
4. Conclusions
The complete cp genome of S. oblata var. alba was determined in this study, which provides theoretical foundation for further study on the phylogenetic relationship of Oleaceae family.
Ethical approval
According to the Regulations of the People’s Republic of China on Wild Plants Protection, S. oblata var. alba is not in the list of national key protection of wild plants. On-site and ex-situ protection of wild plants and scientific research on wild plants are supported in article five of the regulations. No ethical approval or specific permission was needed in this research.
Author contributions
L.X. designed the study. W.X.M., Y.Y.S., and C.B. collected the samples and performed the analyses; H.F.X. and Y.P.X. were involved in the data interpretation; T.G.K. and M.X. participated in the verification and supervision; all authors contributed to the writing and revising of manuscript. All authors finally approved this version.
Supplemental Material
Download JPEG Image (353.9 KB)Supplemental Material
Download JPEG Image (703.7 KB)Supplemental Material
Download JPEG Image (475.2 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. ON920512. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA854692, SRR19960680, and SAMN29446418, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Cheng YQ, Jiang ZM, Cai J. 2021. Characteristic and phylogenetic analyses of chloroplast genome for Syringa komarowii C.K.Schneid. (Oleaceae) from Huoditang, China, an important horticultural plant. Mitochondrial DNA B Resour. 6(4):1521–1522.
- Gao S, Bai JQ, Liu ML, Wang PF, Li N, Yang L, Wang XP, Li ZH. 2019. The complete chloroplast genome of Forsythia mira, an endemic medicinal shrub in China. Mitochondrial DNA B Resour. 5(1):57–58.
- Li JH, Alexander JH, Zhang DL. 2002. Paraphyletic Syringa (Oleaceae): evidence from sequences of nuclear ribosomal DNA ITS and ETS regions. Syst Bot. 27:592–597.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704.
- Liu J, Xiong P, Wang W, Wang W, Zhang W, Qin Y, Wang J. 2020. Characterization of the complete chloroplast genome of Syringa wolfii. Mitochondrial DNA B Resour. 5(3):2051–2053.
- Nemati Z, Harpke D, Gemicioglu A, Kerndorff H, Blattner FR. 2019. Saffron (Crocus sativus) is an autotriploid that evolved in Attica (Greece) from wild Crocus cartwrightianus. Mol Phylogenet Evol. 136:14–20.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Olofsson JK, Cantera I, Van de Paer C, Hong-Wa C, Zedane L, Dunning LT, Alberti A, Christin PA, Besnard G. 2019. Phylogenomics using low-depth whole genome sequencing: a case study with the olive tribe. Mol Ecol Resour. 19(4):877–892.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLOS One. 7(2):e30619.
- Wang L, Wang N, Sun J. 2019. Complete chloroplast genome sequence of Ligustrum quihoui (Oleaceae): genome structure and genomic resources. Mitochondrial DNA B Resour. 4(2):4047–4048.
- Wang J, Wang W, Xiong P, Ye D, Jing M, Zhou H. 2020. The sequence and characterization of the complete plastome of Syringa reticulata subsp. pekinensis (Oleaceae). Mitochondrial DNA B Resour. 5(2):2015–2017.
- Wang W, Yu H, Wang J, Lei W, Gao J, Qiu X, Wang J. 2017. The complete chloroplast genome sequences of the medicinal plant Forsythia suspensa (Oleaceae). Int J Mol Sci. 18(11):2288.
- Wang Y, Lu L, Li J, Li H, You Y, Zang S, Zhang Y, Ye J, Lv Z, Zhang Z, et al. 2022. A chromosome-level genome of Syringa oblata provides new insights into chromosome formation in Oleaceae and evolutionary history of lilacs. Plant J. 111(3):836–848.
- Yi X, Li M, Chen L, Wang X. 2019. The complete chloroplast genome of Fraxinus pennsylvanica (Oleaceae). Mitochondrial DNA B Resour. 4(1):1932–1933.
- Zhang J, Jiang Z, Su H, Zhao H, Cai J. 2019. The complete chloroplast genome sequence of the endangered species Syringa pinnatifolia (Oleaceae). Nord J Bot. 37(5).
- Zhao M, Zhang Y, Xin Z, Meng X, Wang W. 2020. The complete chloroplast genome of Syringa oblata (Oleaceae). Mitochondrial DNA B Resour. 5(3):2278–2279.

