Abstract
Corethrodendron multijugum (Maxim.) (Fabaceae: Corethrodendron), also known as Hedysarum multijugum, is an important medicinal plant and is widely used in traditional Chinese medicine. To better understand the diversity and phylogeny of C. multijugum and other Fabaceae species, we sequenced and annotated the complete chloroplast genome of C. multijugum using the Illumina HiSeq 2500 platform. This complete genome was 122,994 bp long, and encodes a total of 110 genes, including 76 protein-coding genes (PCGs), 30 transfer RNA genes (tRNAs), and four ribosomal RNA unit genes (rRNAs). The C. multijugum plastid with a G + C content of 34.5% presents a negative AT -skew (–0.002) and a positive GC -skew (0.032). Phylogenetic analysis revealed that C. multijugum is more closely related to Hedysarum petrovii. This study provides genetic resource information for the further study of Corethrodendron.
Introduction
Corethrodendron multijugum (Maxim.) (Choi and Ohashi Citation2003), also known as Hedysarum multijugum (Bull Citation1881), is a perennial subshrub or herbaceous plant with woody base (). It is widely distributed in western part of China, grows on gravel slopes and river banks in desert or grassland areas (Wang and Pang Citation1991), and is mostly produced abroad in Mongolia and Russia. This plant has deep roots, strong cold and drought tolerance (Cheng Citation1987; Pan et al. Citation2014; Gu et al. Citation2015). It is not only a widely used traditional Chinese medicine, but also an excellent feed, soil, and water conservation plant, which possesses important medicinal and economic values (Wang and Pang Citation1991; Wang et al. Citation2001, Citation2002; Liu Citation2007; Liu et al. Citation2015). C. multijugum previously belonged to the genus Hedysarum and was recognized as Hedysarum multijugum (Maxim.) (Bull Citation1881). However, the genus Hedysarum was recently revised to the genus Corethrodendron based on morphological and molecular evidence of several barcoding regions, including plastid and nuclear regions (Choi and Ohashi Citation2003). In previous studies, four species of Hedysarum have been identified by ISSR (Sun et al. Citation2017; Cao et al. Citation2021); however, the complete chloroplast genome sequence of C. multijugum has not been reported and its genetic composition and phylogenetic relationships remain unclear.
Figure 1. Morphology and habitat map of Corethrodendron multijugum. (a–c) From Yueliang Bay Park in Guide County, Hainan Tibetan Autonomous Prefecture, Qinghai Province, China (101°43′95″ E, 36°04′61″ N). Photograph by Ying Liu.

In order to better understand the evolution and phylogeny of Corethrodendron, we sequenced the complete chloroplast genome of C. multijugum. This helps provide additional information about the plastid evolution and intrageneric diversity of Corethrodendron, which is crucial for further understanding of the evolution and phylogeny of Corethrodendron.
Materials and methods
The fresh leaves of a single individual were collected from Yueliang Bay Park in Guide County, Hainan Tibetan Autonomous Prefecture, Qinghai Province, China (101°43′95″ E, 36°04′61″ N), in August 2022. After on-site collection, the samples were immediately flash-frozen in liquid nitrogen and then directly mailed to the sequencing company (Biotechnologies Inc., Shanghai, China). The specimens (voucher no. QHGN-YLW22B) have been deposited in the Key Laboratory of Superior Forage Germplasm in the Qinghai-Tibetan Plateau. College of Qinghai Academy of Animal and Veterinary Sciences, Qinghai University, Xining, China (URL: https://mky.qhu.edu.cn/index.htm, contact: Ying Liu [email protected]).
Total genomic DNA was extracted from a single specimen using a DNeasy Tissue Kit (Qiagen, Inc., Hilden, Germany). The total read length of raw data was 24,784,146 bp. The complete plastome was assembled as follows: the total genomic DNA was extracted from leaf tissues using the TruSeq DNA sample Preparation kit (Vazyme, Nanjing, China), and then sequenced using the Illumina Hiseq 2500 platform (Illumina, San Diego, CA) with paired-end reads of 150 bp by Genesky Biotechnologies Inc. (Shanghai, China). After trimming, the high-quality paired-end reads were assembled through metaSPAdes software (version 3.13.0) (Korobeynikov Citation2017). The sequence was matched to the genome, and all positions of the read-mapping depth of the assembled genome were statistically obtained (Figure S1). The complete chloroplast genome of Hedysarum petrovii (GenBank accession MT120797) was used as the reference genome, and genome annotation was performed using the program CPGAVAS2 (version N/A) (Shi et al. Citation2019) to compare the sequences with the chloroplast genome of H. petrovii. We then corrected the annotations by comparing them with the published complete chloroplast genomes of Hedysarum species using Geneious R8 (Biomatters Ltd, Auckland, New Zealand). A plastome map was constructed using the CPGView software (Liu et al. Citation2023) (http://www.1kmpg.cn/cpgview). Finally, the assembled chloroplast genome and its detailed annotations were submitted to GenBank under the accession number NC069301.
Results and discussion
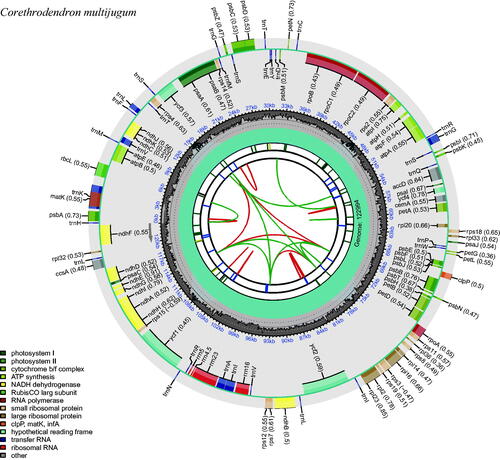
We obtained the complete chloroplast genome of C. multijugum as a circular DNA molecule () of 122,994 bp long. Different from typical quadripartite structure of most angiosperm chloroplast genomes, the chloroplast genome of C. multijugum does not have the typical quadripartite structure consisting of a large single-copy (LSC), a small single-copy (SSC), and a pair of inverted repeats (IRs), the similar result is also demonstrated for Hedysarum polybotrys var. alaschanicum (Cao et al. Citation2021) and Astragalus complanatus (Yang Citation2021). The base compositions of the chloroplast genome were uneven (32.7% A, 32.8% T, 16.7% C, and 17.8% G). The overall GC and AT content of the whole genome is 34.5% and 63.5%, the genome presenting a negative AT-skew (–0.002) and a positive GC-skew (0.032) on the J-strand. The complete chloroplast genome encodes 110 genes, including 76 protein-coding genes (PCGs), 30 transfer RNA genes (tRNAs), and four ribosomal RNA unit genes (rRNAs). Among these genes, 15 genes (atpF, ndhA, ndhB, petB, petD, rpI2, rps12, rpI16, rpoC1, trnA-UGC, trnG-GCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) had one intron, and two (ycf2 and ycf3) contained two introns. A total of 65 SSR markers ranging from mononucleotide to tetranucleotide repeat motifs were identified in the C. multijugum chloroplast genome, mononucleotide had the most repeats.
Figure 2. The plastome genome map of Corethrodendron multijugum under this study. From the inner circle, the first circle depicts distributed repeats connected by red (forward direction) and green (reverse direction) arcs, respectively. The following circle displays tandem repeats denoted by short blue bars. The sequences of microsatellites are depicted as short green bars. The fourth circle displays the sizes of LCS, SSC, and IR (this plastid lacks annotation of these three regions). The fifth circle illustrates the distribution of GC contents along the plastome (dark grey: GC contents, light grey: background). The sixth circle displays the genes with colored boxes. The outer and inner colored boxes present transcribed clockwise and counterclockwise genes, respectively.
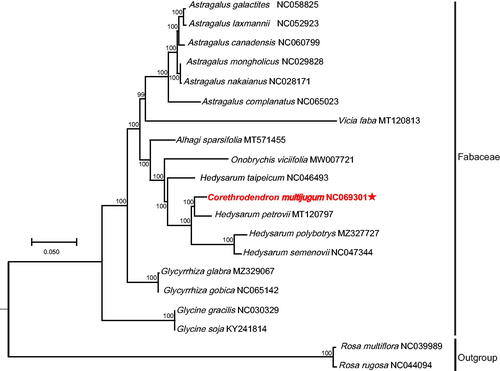
A phylogenetic tree was reconstructed to confirm the phylogenetic of C. multijugum. At present, only the four species chloroplast genome data of Hedysarum polybotrys, H. taipeicum, H. semenovii, and H. petrovii have been published and are available in the NCBI database. All of the 20 chloroplast genome sequences were obtained from GenBank and used for phylogenetic analysis, contain the complete chloroplast sequences of C. multijugum, and the sequences of other seventeen species in seven genera (four Hedysarum species, one Vicia species, two Glycine species, six Astragalus species, two Glycyrrhiza species, one Onobrychis species, and one Alhagi species) of Fabaceae. Two Rosa species (Rosa rugosa and R. multiflora) of Rosaceae were used as outgroups. All of the 20 chloroplast genome sequences aligned using MEGA7 (Kumar et al. Citation2016) with default parameters. The maximum-likelihood (ML) tree was built using MEGA7 with the bootstrap set to 1000. Phylogenetic analysis indicated a strong sister relationship between C. multijugum and H. petrovii ().
Figure 3. Chloroplast phylogeny of 18 Fabaceae species based on the complete chloroplast genome sequences. The asterisk represents the assembled plastome sequence in this study. The clades of species are represented with black lines. The following sequences of each species were used: Astragalus canadensis NC060799 (unpublished), A. complanatus NC065023 (unpublished), A. galactites NC058825 (Ding et al. Citation2021), A. laxmannii NC052923 (Liu et al. Citation2020), A. mongholicus NC029828 (Lei et al. Citation2016), A. nakaianus NC028171 (Choi et al. Citation2016), Glycine gracilis NC030329 (Gao and Gao Citation2017), G. soja KY241814 (unpublished), Glycyrrhiza glabra MZ329067 (unpublished), G. gobica NC065142 (unpublished), Corethrodendron multijugum NC069301 (this study), Hedysarum petrovii MT120797 (unpublished), H. polybotrys MZ327727 (Cao et al. Citation2021), H. semenovii NC047344 (Zhang et al. Citation2020), H. taipeicum NC046493 (She et al. Citation2019), Vicia faba MT120813 (unpublished), Onobrychis viciifolia MW007721 (Fu et al. Citation2021), Alhagi sparsifolia MT571455 (Wang et al. Citation2020), Rosa multiflora NC039989, and R. rugosa NC044094 (Kim et al. Citation2019). Unpublished in the legend indicates that the citations have not been published.
In the present study, the chloroplast genome of C. multijugum is reported for the first time. We described the sequence structures and annotated genes in the genome. Its sequence length was found to be 122,994 bp, similar to that of other Corethrodendron (Hedysarum) species. What is different from other studies is that compared with other previously published chloroplast genomes of Corethrodendron (Hedysarum) species, the C. multijugum chloroplast genome does not have the typical quadripartite structure, and similar genetic structure results have been demonstrated in both genus Hedysarum (Cao et al. Citation2021) and genus Astragalus (Yang Citation2021).What is special about this study is that the phylogenetic tree was reconstructed to confirm the phylogenetic of C. multijugum for the first time. The chloroplast genome of C. multijugum will contribute to a better understanding of the evolutionary mode of the chloroplast genome and provide more evidence for the identification and application of Corethrodendron species.
Author contributions
Ying Liu (corresponding author) collected the materials, conception and designed the experiment. Li-Jun Zhang (first author) analyzed and interpretation of the data, and drafting of the manuscript. After that, Ying Liu and Jian-Jun Shi revised it critically for intellectual content of this article and agreed to the final approval to be published. All authors reviewed the manuscript and agreed to be accountable for all aspects of the work.
Ethical approval
Based on No. 221(E) Article 15: (1)-ii) of the International Union for the Protection of New Varieties of Plants (UPOV) in 1991, this study can be conducted without ethical approval or permission. Research on plant chloroplast genome sequencing does not require ethical approval.
Supplemental Material
Download PDF (305.7 KB)Supplemental Material
Download PDF (1.4 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank database of NCBI at https://www.ncbi.nlm.nih.gov/ under accession no. NC069301 (https://www.ncbi.nlm.nih.gov/nuccore/NC_069301.1). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA896184 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA896184), SRA: SRR22200104 (https://www.ncbi.nlm.nih.gov/sra/?term=SRR22200104), and SAMN31536048 (https://www.ncbi.nlm.nih.gov/biosample/?term=SAMN31536048), respectively.
Additional information
Funding
References
- Bull. 1881. Hedysarum multijugum Maxim. Acad Imp Sci Saint-Pétersbourg. 27(4):464.
- Cao JN, Han C, Yang Y. 2021. Characterization of the complete chloroplast genome of Hedysarum polybotrys var. alaschanicum (Fabaceae) and its phylogeny. Mitochondrial DNA B Resour. 6(11):3312–3313.
- Cheng JM. 1987. The excellent herbage of controlling green sand appearance – Hedysarum multijugum. China’s Grasslands. 10(1):39–42.
- Choi BH, Ohashi H. 2003. Generic criteria and an infrageneric system for Hedysarum and related genera (Papilionoideae-Leguminosae). Taxon. 52(3):567–576.
- Choi IS, Kim JH, Choi BH. 2016. Complete plastid genome of Astragalus mongholicus var. nakaianus (Fabaceae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(4):2838–2839.
- Ding X, Zhang C, Gao Y, Bei Z, Yan X. 2021. Characterization of the complete chloroplast genome of Astragalus galactites (Fabaceae). Mitochondrial DNA B Resour. 6(11):3278–3279.
- Fu X, Ji X, Wang B, Duan L. 2021. The complete chloroplast genome of leguminous forage Onobrychis viciifolia. Mitochondrial DNA B Resour. 6(3):898–899.
- Gao CW, Gao LZ. 2017. The complete chloroplast genome sequence of semi-wild soybean, Glycine gracilis (Fabales: Fabaceae). Conserv Genet Resour. 9(2):343–345.
- Gu WY, Wang DX, Liu XL. 2015. Effects of drought stress on physiological indices of resistance of four wild groundcover species. North Hortic. 12(18):81–83.
- Kim Y, Heo KI, Nam S, Xi H, Lee S, Park J. 2019. The complete chloroplast genome of candidate new species from Rosa rugosa in Korea (Rosaceae). Mitochondrial DNA B Resour. 4(2):2433–2435.
- Korobeynikov A. 2017. SPAdes: is there anything new we could develop? Genome Research. 27: 824-834. San Diego, USA.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lei W, Ni D, Wang Y, Shao J, Wang X, Yang D, Wang J, Chen H, Liu C. 2016. Intraspecific and heteroplasmic variations, gene losses and inversions in the chloroplast genome of Astragalus membranaceus. Sci Rep. 6:21669.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704.
- Liu XL. 2007. Excellent wild ecotype ornamental ground cover – Hedysarum multijugum. North Hortic. 1(7):154–155.
- Liu Y, Chen Y, Fu X. 2020. The complete chloroplast genome sequence of medicinal plant: Astragalus laxmannii (Fabaceae). Mitochondrial DNA Part B. 5(3):3643–3644.
- Liu Y, Wang W, Zhao YL, Chen H, Liang H, Zhang Q. 2015. Chemical analysis of the Hedysarum multijugum root by HPLC fingerprinting. J Chin Pharm Sci. 24(10):654–659.
- Pan X, Qiu Q, Li JY, Junhui W, He Q, Su Y, Ma JW, Du K. 2014. Physiological indexes of six plant species from the Tibetan plateau under drought stress. Acta Ecol Sin. 34(13):3558–3567.
- She RX, Li WQ, Xie XM, Gao XX, Wang L, Liu PL, Yan F, Khan H, Ullah I, Zhao P, et al. 2019. The complete chloroplast genome sequence of a threatened perennial herb species Taibai sweetvetch (Hedysarum taipeicum K.T. Fu). Mitochondrial DNA Part B. 4(1):1439–1440.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Sun SY, Wu YX, Chen GL, Zhao YX. 2017. Identification of Astragali Radix and its substitutes by ISSR markers. Mol Plant Breed. 15(1):223–229.
- Wang AH, Deng SW, Duan L, Chen HF. 2020. The complete chloroplast genome of desert spiny semi-shrub Alhagi sparsifolia (Fabaceae) from Central Asia. Mitochondrial DNA B Resour. 5(3):3098–3099.
- Wang KB, Pang XM. 1991. Hedysarum multijugum Maxim. Soil Water Conserv China. 1(3):42.
- Wang W, Chen HB, Wang WM, Zhao YY. 2002. Studies on chemical constituents and biological activities of Hedysarum multijugum. Beijing, China: Peking University.
- Wang W, Chen W, Chen HB, Liu J, Zhao Y. 2001. Studies on chemical constituents of Hedysarum multijugum. J Peking Univ (Health Sci). 33(3):205–208.
- Yang J. 2021. Characterization of the complete plastid genome of the perennial herb Astragalus complanatus Bunge (Fabales: Fabaceae). Mitochondrial DNA B Resour. 6(12):3440–3442.
- Zhang R, Wang YH, Jin JJ, Stull GW, Bruneau A, Cardoso D, De Queiroz LP, Moore MJ, Zhang SD, Chen SY, et al. 2020. Exploration of plastid phylogenomic conflict yields new insights into the deep relationships of Leguminosae. Syst Biol. 69(4):613–622.
