Abstract
Cenchrus alopecuroides Thunb. 1794 is a warm-season ornamental grass with glossy foliage and showy inflorescence, which is widely distributed from eastern Asia to Australia. Although this species is of great economic importance, little genomic sequence data are available. Here, we report the complete chloroplast genome sequence of C. alopecuroides, which provides valuable plastid genomic resources for further studies on this ornamental grass. The chloroplast genome of C. alopecuroides was 138,053 bp in length and exhibited a typical quadripartite structure, which comprised a pair of inverted repeat regions (22,331 bp) separated by a large (81,177 bp) and a small single copy (12,214 bp) region. In total, 110 unique genes were annotated in the chloroplast genome, containing 76 protein-coding genes, 30 transfer RNAs and four ribosomal RNAs. The overall GC content of the chloroplast genome was 38.6%. Phylogenetic analysis based on 11 whole-chloroplast genome sequences of Cenchrus species suggested that C. alopecuroides and C. compressus were sisters to each other and joint sisters to C. clandestinus.
1. Introduction
Cenchrus alopecuroides Thunb. 1794, formerly known as Pennisetum alopecuroides, belongs to the Poaceae family and is widely distributed from eastern Asia to Australia (Thomas and Taylor Citation2021). This species is an attractive and widely popular, moderately sized ornamental grass () that forms dense clusters of narrow leaves and tall, upright, foxtail-shaped inflorescences (Thomas and Taylor Citation2021). Since it produces long flower spikes that erupt and curve out from the plant like a fountain of water, C. alopecuroides is also called swamp foxtail grass and/or foxtail fountain grass (Toon et al. Citation2018). Currently, many cultivars of this species are commercially available for horticulture practices, with a wide range of plant height (0.30–1.52 m), various flower colors (e.g. purple, pink and white), and autumn foliage (http://www.missouribotanicalgarden.org). Despite its high economic importance, there is little genomic sequence data available for C. alopecuroides. Here, we report the chloroplast genome sequence of C. alopecuroides and reconstruct its phylogenetic relationship with other Cenchrus species. The main objective of this study was to provide a basis for further research and utilization of this economically important species.
2. Materials and methods
2.1. Plant material, DNA extraction, and genome sequencing
Fresh leaves of C. alopecuroides (, taken by the author Zhiyong Wang) were sampled from the Xinyang Agricultural and Forestry University (E114°12′, N32°16′). A voucher specimen was deposited at the College of Horticulture, Xinyang Agricultural and Forestry University (https://www.xyafu.edu.cn/en/, contact Peiling Li, [email protected]) under voucher number Cal20211202. DNA was extracted using DNA Plantzol Reagent (Invitrogen Trading Co., Ltd, Shanghai, China). DNA library construction and 150 bp paired-end sequencing were performed on the Illumina HiSeq4000 platform, and approximately 6 GB of raw data was obtained. Library construction and sequencing were conducted by Novogene Bioinformatics Technology Co., Ltd. (Beijing, China).
2.2. Chloroplast genome assembly and annotation
The chloroplast genome was assembled using the GetOrganelle pipeline (Jin et al. Citation2020), with the suggested default parameters. To evaluate the quality and completeness of the genome assembly, the genome coverage was estimated by mapping all the clean reads back to the chloroplast genome sequence, using CLC Genomics Workbench 23.0.1 (https://digitalinsights.qiagen.com). The assembled plastome sequence was annotated using the PGA (Plastid Genome Annotator) program (Qu et al. Citation2019), with the annotated chloroplast genome sequence of C. americanus (MN180104) as a reference. The exon/intron boundaries and the start and stop codons were further verified by comparison with the reference genome using Geneious Prime® 2022.0.1. software (http://www.geneious.com). The annotated chloroplast genome sequence of C. alopecuroides has been deposited in GenBank (accession number: ON206984). A chloroplast genome map was drawn using CPGview (http://www.1kmpg.cn/cpgview) (Liu et al. Citation2023).
2.3. Phylogenetic analyses
Phylogenetic analyses were conducted on C. alopecuroides and 10 Cenchrus species, with Stenotaphrum secundatum (KY432796) and Setaria italica (MK348605) as outgroups. The chloroplast genomes of these 13 species were aligned in Geneious Prime® 2022.0.1 using the MAFFT v.7 plugin (Katoh and Standley Citation2013). The best nucleotide substitution model was determined using jModelTest v2.1.4 (Posada Citation2008) under the Akaike Information Criterion (AIC) criterion, and the GTR + G + I substitution model was selected. Maximum likelihood (ML) analysis was carried out using RAxML-HPC v.8.2.8 (Stamatakis Citation2014) on the CIPRES Science Gateway (http://www.phylo.org/portal2/). Clade support values were derived from 1,000 bootstrap replicates.
3. Results and discussion
3.1. Chloroplast genome features
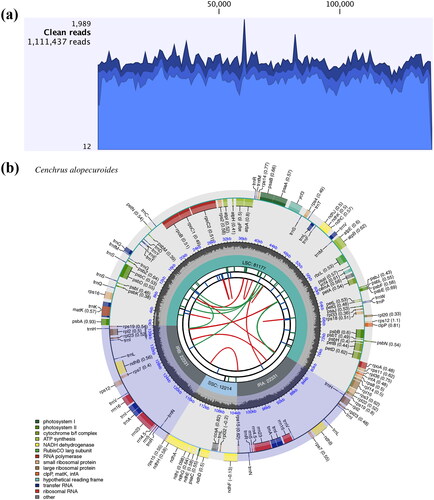
A total of 1,111,437 clean reads representing 2.5% of the total whole genome sequencing data were mapped to the chloroplast genome sequence of C. alopecuroides, with an average coverage depth of 1207.6× (). The chloroplast genome sequence of C. alopecuroides was 138,053 bp in length, and exhibited a typical quadripartite structure consisting of a pair of inverted repeat regions (IRs) of 22,331 bp separated by a large single-copy (LSC) region of 81,177 bp and a small single copy (SSC) region of 12,214 bp (). The chloroplast genome encoded 130 genes, of which 110 (76 protein-coding genes, 30 tRNA genes, and 4 rRNA genes) were unique and 20 (8 protein-coding genes, 8 tRNA genes, and 4 rRNA genes) were duplicated in the IRs (). Eight protein-coding genes (rps16, atpF, petB, petD, rpl16, rpl2, ndhB, and ndhA) and six tRNA genes (trnK-UUU, trnG-UCC, trnL-UAA, trnV-UAC, trnI-GAU, and trnA-UGC) contained a single intron, whereas three genes (ycf3, rps12, and clpP) contained two introns. The structure of cis-/trans-splice genes were shown in Figure S1. The overall GC content was 38.6%, whereas the CC content in the IR, LSC, and SSC were 44.1, 36.5, and 33.2%, respectively.
Figure 2. The chloroplast genome coverage depth and map of Cenchrus alopecuroides. (a) The mapped read depth of Cenchrus alopecuroides chloroplast genome. (b) The chloroplast genome map of Cenchrus alopecuroides. The map contains six circles. From the center to outward, the first circle shows the distributed repeats connected with red (forward) and green (reverse) arcs. The second circle shows the tandem repeats as short bars. The third circle shows the microsatellite sequences marked with short bars. The fourth circle shows the size of the LSC, SSC, and IRs. The fifth circle shows the GC contents along the plastome. The sixth circle shows the genes having different colors based on their functional groups. Genes shown outside the circle are transcribed clockwise, and those inside counter-clockwise.
3.2. Phylogenetic analyses
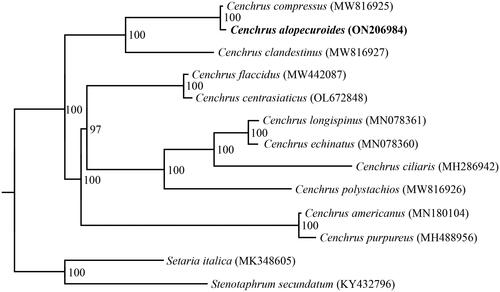
Comparative chloroplast genome analyses of C. alopecuroides and 10 previously reported chloroplast genomes of Cenchrus showed that the chloroplast genomes within this genus were conserved in terms of genome size, genome structure, and gene content (e.g. Bhatt and Thaker Citation2018; Wu and Zhou Citation2019; Zhang et al. Citation2021). The phylogenetic results using 100% bootstrap values for almost all nodes, strongly supported the monophyly of Cenchrus, and also indicated that C. alopecuroides and C. compressus formed a sister group, which was further sister to C. clandestinus ().
Figure 3. Phylogenetic tree inferred by the maximum likelihood (ML) method based on complete chloroplast genomes of 11 Cenchrus species, with Stenotaphrum secundatum (KY432796) and Setaria italica (MK348605) as outgroups. Numbers near the nodes represent ML bootstrap values. The following sequences were used: MW816925–MW816927 (Xu et al. Citation2021), MW442087 (Liu, W., Direct submission), OL672848 (Zhang, L., Direct submission), MN078360–MN078361 (Hyun et al. Citation2019), MH286942 (Bhatt and Thaker Citation2018), MN18014 (Xu et al. Citation2019), MN488956 (Bhatt and Thaker Citation2018), MK348605 (Liu et al. Citation2019), KY432796 (Gallaher et al. Unpublished).
4. Conclusion
In this study, the chloroplast genome of C. alopecuroides was reported for the first time, and its organization was described. The complete chloroplast genome was 138,053 bp in length and encoded a total of 130 genes. Comparative chloroplast genome analyses revealed that the chloroplast genomes within this genus were conserved in terms of genome size and structure, and gene content. Phylogenetic analysis of the Cenchrus species suggested that C. alopecuroides and C. compressus are sisters to each other and joint sisters to C. clandestinus. The genomic resources presented here will not only offer valuable information for further genetic and evolutionary studies on C. clandestinus but will also be useful for breeding, cultivation and utilization of this economically important species.
Ethical approval
We confirm that all the research meets ethical guidelines and adheres to the legal requirements of the Xinyang Agricultural and Forestry University (China).
Author contributions
ZW and PL designed the project. LY, JY and YT collected samples and analyzed data. ZW and PL wrote the manuscript. JY and PL revised the manuscript. All authors read and approved the manuscript. All the authors meet the criteria for authorship as per the ICMJE criteria.
Supplemental Material
Download TIFF Image (321.6 KB)Disclosure statement
No potential conflict of interest was reported by the author(s). The study was permitted under permission from Xinyang Agricultural and Forestry University.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. ON206984. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA871033, SRR21166642, and SAMN30399920, respectively.
Additional information
Funding
References
- Bhatt P, Thaker V. 2018. The complete chloroplast genome of Cenchrus ciliaris (Poaceae). Mitochondrial DNA B Resour. 3(2):674–675.
- Hyun J, Jung J, Do HDK, Kim JH. 2019. Preliminary research for molecular markers of two invasive toxic weeds, Cenchrus L. (Poaceae) species, based on NGS technique. Proceedings of the Plant Resources Society of Korea Conference; The Plant Resources Society of Korea. p. 71.
- Jin J, Yu W, Yang J, Song Y, DePamphilis C, Yi T, Li D. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley D. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Liu D, Cui Y, Li S, Bai G, Li Q, Zhao Z, Liang D, Wang C, Wang J, Shi X, et al. 2019. A new chloroplast DNA extraction protocol significantly improves the chloroplast genome sequence quality of foxtail millet (Setaria italica (L.) P. Beauv.). Sci Rep. 9(1):1–9.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704.
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25(7):1253–1256.
- Qu X, Moore M, Li D, Yi T. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Thomas J, Taylor M. 2021. Evaluation of chemical control methods of fountain grass. hortte. 31(4):382–384.
- Toon A, Sacre E, Fensham RJ, Cook LG. 2018. Immigrant and native? The case of the swamp foxtail Cenchrus purpurascens in Australia. Divers Distrib. 24(8):1169–1181.
- Wu Y, Zhou H. 2019. The complete chloroplast genome sequence of Cenchrus purpureus. Mitochondrial DNA B Resour. 4(1):51–52.
- Xu J, Liu C, Song Y, Li M. 2021. Comparative analysis of the chloroplast genome for four Pennisetum species: molecular structure and phylogenetic relationships. Front Genet. 12:687844.
- Xu J, Song Y, Jing X, Li M. 2019. Characterization of the complete chloroplast genome sequence of Pennisetum glaucum and its phylogenetic implications. Mitochondrial DNA B Resour. 4(2):3764–3765.
- Zhang L, Liu D, Ma Y, Yang S, Wang X, Wang Y, Dong R. 2021. The complete chloroplast genome sequence of Pennisetum centrasiaticum, a widespread grass in Tibet, China. Mitochondrial DNA B Resour. 6(8):2105–2106.

