Abstract
Lithocarpus litseifolius (Hance) Chun 1837 is an evergreen tree of Fagaceae, which can be used as sweet tea, natural sweetener, and precious medicinal material. The complete chloroplast genome of L. litseifolius was sequenced and its phylogenetic relationship was analyzed in this study. The chloroplast genome of L. litseifolius has a circular structure with a length of 161,322 bp, and it contains a pair of inverted repeat regions (IRs 25,897 bp), a large single copy (LSC 90,551 bp), and a small single copy (SSC 18,977 bp). There were 131 genes identified, including 37 tRNA, 8 rRNA, and 86 mRNA genes. Phylogenetic analysis of 23 species of Fagaceae indicated that Lithocarpus is monophyletic with strong bootstrap, and L. litseifolius is genetically closely related to Lithocarpus polystachyus.
1. Introduction
Lithocarpus litseifolius (Hance) Chun 1837 is an evergreen tree in the Fagaceae family (Cheng et al. Citation2016). The tender, containing kinds of sweet materials, has been accepted as sweet tea for thousands of years (Liu et al. Citation2021). More recently, L. litseifolius is regarded as a precious Chinese medicine for its unique effect in preventing diabetes (Wang et al. Citation2021). L. litseifolius has been approved as a new food raw material resource, which has great development value (Kalleli et al. Citation2019). The complete chloroplast genome of L. litseifolius has not been sequenced, so it is meaningful to sequence the chloroplast genome to study its evolutionary relationship. Here, we assembled and annotated the complete chloroplast genome of L. litseifolius. And the evolutionary relationships between different genera in Fagaceae were studied.
2. Materials and methods
The plant of L. litseifolius was shown in . Fresh leaf materials of L. litseifolius were collected from Liangyaping town, Xupu county, China (27°48′7.65″N, 110°35′6.73″E). A voucher specimen was identified by Shengen Xiao and deposited at Hunan Yao Tea Engineering Technology Center (http://www.oncolbio.com/) with the voucher number H1-2-001 under the charge of Yuqiao Tian ([email protected]). Fresh leaves were used to extract whole genomic DNA using the modified CTAB method (Yang et al. Citation2014), and the detailed extraction steps were placed in the supplementary material. Genomic DNA was fragmented into 350 bp fragments by an ultrasonic processor, and Agilent 2100 was used to construct the whole-genome DNA sequencing library, which was then sequenced on an Illumina NovaSeq 6000 platform. About 4.77 Gb clean data was obtained by using the fastp (version 0.20.0; https://github.com/opengene/fastp) (Table S1). Subsequently, SPAdes was used to assemble the complete chloroplast genome (http://cab.spbu.ru/software/spades/) with the chloroplast genome of Lithocarpus hancei (MW375417.1) as a reference (Ma et al. Citation2021). Circos (version 0.69-9) was used to map the genome sequencing data (Figure S1). The schematic map of the cis-splicing genes was shown in Figure S2, and the schematic map of the trans-splicing gene rps12 was shown in Figure S3. Finally, the complete chloroplast genome sequence of L. litseifolius was annotated in Prodigal (https://www.github.com/hyattpd/Prodigal), which was deposited into NCBI with an accession number OM048987.1.
Figure 1. The reference image of L. litseifolius. Trees to 20 m tall. Leaf blade elliptic, obovate-elliptic, ovate, or rarely narrowly elliptic, 8–18 × 3–8 cm, papery to sub leathery, margin entire, apex acuminate to acute. Its tender leaves are sweet, commonly known as sweet tea. Female and androgynous inflorescences are usually 2–6 congested at apex of branches, spiciform, to 35 cm; cupules in clusters of 3–5. This image was taken by Xiaoyan Qiu in Liangyaping Town, China (27°48′7.65″N, 110°35′6.73″E).

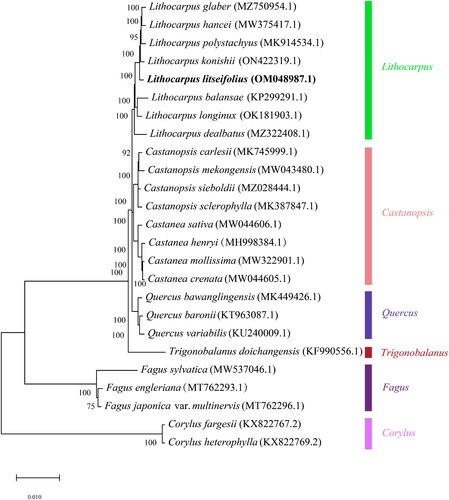
To infer the phylogenetic relationship of L. litseifolius in Fagaceae, the complete chloroplast genome sequences of 23 species in this family were used for phylogenetic analysis, with Corylus fargesii (KX822767.2) and Corylus heterophylla (KX822769.2) as outgroups. All the complete chloroplast genome sequences of these species were downloaded from GenBank and the accession numbers were shown in . These sequences were adjusted to the same starting point and after being aligned by MAFFT, the Maximum Likelihood (ML) tree was obtained by using RaxML with a GTR + GAMMA model and 1000 replicates for a bootstrap test (Stamatakis Citation2014).
Figure 2. Maximum-likelihood phylogenetic tree for L.litseifolius was constructed based on 25 complete chloroplast genomes using Corylus fargesii and Corylus heterophylly as outgroups. The following sequence was used: Lithocarpus hancei MW375417.1 (Ma et al. Citation2021), Lithocarpus polystachyus MK914534.1 (Li et al. Citation2019), Lithocarpus konishii ON422319.1 (unpublished), Lithocarpus litseifollus OM048987.1 (unpublished), Lithocarpus balansae KP299291.1 (Li et al. Citation2018), Lithocarpus longinux OK181903.1 (Wu et al. Citation2022), Lithocarpus dealbatus MZ322408.1 (Shelke et al. Citation2022), Castanopsis carlesii MK745999.1 (Liu et al. Citation2019), Castanopsis mekongensis MW043480.1 (unpublished), Castanopsis sieboldii MZ028444.1 (Park et al. Citation2021), Castanopsis sclerophylla MK387847.1 (Shelke et al. Citation2022), Castanea sativa MW044606.1 (unpublished), Castanea henryi MH998384.1 (Gao et al. Citation2019), Castanea mollissima MW322901.1 (Zhang et al. Citation2021), Castanea crenata MW044605.1 (Jeong et al. Citation2019), Quercus bawanglingensis MK449426.1 (Ma et al. Citation2021), Quercus haronii KT963087.1 (Ma et al. Citation2021), Quercus variabilis KU240009.1 (Li et al. Citation2018), Trigonobalanus doichangensis KF990556.1 (Park and Oh Citation2020), Fagus sylvatica MW537046.1 (Mishra et al. Citation2021), Fagus engleriana MT762293.1 (unpublished), Fagus japonica MT762296.1 (unpublished), Corylus fargesii KX822767.2 (Hu et al. Citation2017), and Corylus heterophylla KX822769.2 (Ma et al. Citation2021).
3. Results
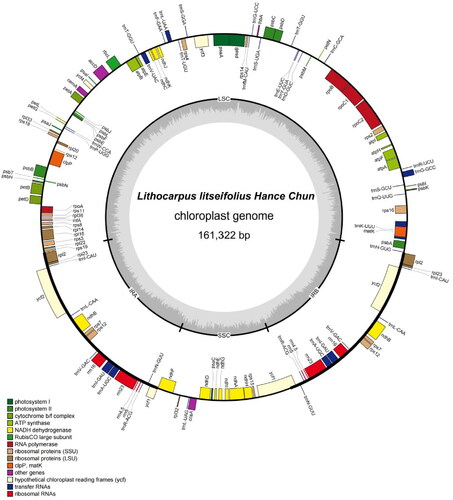
The chloroplast genome of L. litseifolius has a circular structure with a length of 161,322 bp, and it contains a pair of inverted repeat regions (IRs 25,897 bp), a large single copy (LSC 90,551 bp), and a small single copy (SSC 18,977 bp) (). There were 131 genes identified, including 37 tRNA, 8 rRNA, and 86 mRNA genes. Among the annotated genes, nine protein-coding genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpl2, rpoC1, rps16) and six transfer RNA genes (trnA-UGC, trnG-GCC, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC) contained one intron. Besides, three protein-coding genes (ycf3, clpP, and rps12) had two introns. Total GC content is 36.32%, and the corresponding values of the IR, LSC, and SSC, were 30.71%, 34.57%, and 30.71%, respectively. As shown in the phylogenetic tree (), all of the nodes in the phylogenetic trees had high bootstrap support values. L. litseifolius belonged to a unique species in Lithocarpus genus compared to other plants in the same family, this result was consistent with plant taxonomy, and L. litseifolius was genetically closely related to Lithocarpus polystachyus.
Figure 3. The circular map of L. litseifolius chloroplast genome. Genes with different functions are shown in different colors. Genes shown on the outside and inside of the circle are transcribed clockwise and counterclockwise, respectively. The grey circle inside represents the GC content. The SSC and LSC regions are separated by IRs (IRA and IRB).
4. Discussion and conclusion
There are about 900 species in Fagaceae, which are divided into seven genera, Castanea, Castanopsis, Cyclobalanopsis, Fagus, Lithocarpus, Quercus, and Trigonobalanus. And Lithocarpus is the second-largest genus with about 330 species (Shelke et al. Citation2022). The phylogeny of the Fagaceae family has received a lot of attention. The use of a few nuclear markers such as ITS1 and ITS2 or their combinations by earlier workers was unable to distinguish species clearly when used to infer the phylogenetic relationship in Fagaceae (Pang et al. Citation2019). In this study, the complete chloroplast genome of L. litseifolius was sequenced and its phylogenetic relationship was analyzed. Phylogenetic analysis of 23 species of Fagaceae strongly supported that Lithocarpus was monophyletic with strong bootstrap and L. litseifolius was genetically closely related to Lithocarpus polystachyus. This phylogenetic result was similar to those of Li et al. (Citation2019), Ma et al. (Citation2021), and Wu et al. (Citation2022). These results probably implied that the chloroplast genomes can better resolve the inter-specific relationship within Fagaceae. Further study into evolutionary biology, population genetics, and species identification of L. litseifolius with related species might be built upon the entire structure of the chloroplast genome shown here.
Authors’ contributions
Xiaoyan Qiu performed experiments and drafted the manuscript, Shengen Xiao conceived and designed the study and directed the writing, Yuqiao Tian and Ziqiang Li guided data analysis and interpretation. All authors approved the final version of the manuscript.
Ethical approval
The sample (L. litseifolius) was collected strictly by guidelines provided following the Hunan Agriculture University, and Chinese regulations. No ethical approval is required in this study.
Supplemental Material
Download MS Word (6.7 MB)Supplemental Material
Download PNG Image (306.7 KB)Supplemental Material
Download PNG Image (501.1 KB)Supplemental Material
Download PNG Image (5.8 MB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession number OM048987.1. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA792474, SRR17333522, and SAMN24425764, respectively.
Additional information
Funding
References
- Cheng J, Lyu LS, Shen YB, Li KX, Liu ZH, Wang WX, Xie L. 2016. Population structure and genetic diversity of Lithocarpus litseifolius (Fagaceae) assessed using microsatellite markers. Nord J Bot. 000:001–009.
- Gao XX, Yan F, Liu M, Zulfiqar S, Zhao P. 2019. The complete chloroplast genome sequence of an endemic species Pearl chestnut (Castanea henryi). Mitochondrial DNA Part B. 4(1):551–552.
- Hu GL, Cheng LL, Lan YP, Cao QC, Wang XQ, Huang WG. 2017. The complete chloroplast genome sequence of the endangered Chinese endemic tree Corylus fargesii. Conservation Genet Resour. 9(2):225–227.
- Jeong KM, Dong KT, Sang L, Ryeon LH, Chulwoo K, Hyoshin L, Jun PE. 2019. The complete chloroplast genome of Castanea crenata Sieb. & Zucc. Mitochondrial DNA Part B. 4(2):3864–3865.
- Kalleli F, Bettaieb RI, Wannes WA, Boughalleb F, Hammami M, Saidani TM, M'Hamdi M. 2019. Chemical composition and antioxidant potential of essential oil and methanol extract from Tunisian and French fennel (Foeniculum vulgare Mill.) seeds. J Food Biochem. 43(8):e12935.
- Liu Y, Liu HY, Xia Y, Guo H, He XQ, Li H, Wu DT, Geng F, Lin FJ, Li HB, et al. 2021. Screening and process optimization of ultrasound assisted extraction of main antioxidants from sweet tea (Lithocarpus litseifolius [Hance] Chun). Food Bio Sci. 43:101277.
- Li YQ, Guo W, He P, Yu LH. 2019. The complete chloroplast genome of sweet tea (Lithocarpus polystachyus). Mitochondrial DNA B Resour. 4(2):2489–2490.
- Liu B, Zeng QM, Jiang YT, Lin RQ, Liu ZJ, Chen SP. 2019. The complete chloroplast genome sequence of Castanopsis carlesii (Fagaceae). Mitochondrial DNA Part B. 4(1):2076–2077.
- Li X, Li YF, Zang MY, Li MZ, Fang YM. 2018. Complete chloroplast genome sequence and phylogenetic analysis of Quercus acutissima. IJMS. 19(8):2443.
- Ma CX, Yan HF, Ge XJ. 2021. The complete chloroplast genome of Lithocarpus hancei (Benth.) Rehd (Fagaceae) from Zhejiang, China. Mitochondrial DNA B Resour. 6(7):2022–2023.
- Mishra B, Ulaszewski B, Ploch S, Burczyk J, Thines M. 2021. A circular chloroplast genome of Fagus sylvatica reveals high conservation between two individuals from Germany and one individual from Poland and an alternate direction of the small single-copy region. Forests. 12(2):180.
- Park JS, Xi H, Son J, Shin HT, Kang H, Park S. 2021. The complete chloroplast genome of Castanopsis sieboldii (Makino) Hatus (Fagaceae). Mitochondrial DNA B Resour. 6(9):2743–2745.
- Pang X, Liu H, Wu S, Yuan Y, Li H, Dong J, Liu Z, An C, Su Z, Li B. 2019. Species identification of Oaks (Quercus L., Fagaceae) from gene to genome. IJMS. 20(23):5940.
- Park JS, Oh SH. 2020. A second complete chloroplast genome sequence of Fagus multinervis Nakai (Fagaceae): intraspecific variations on chloroplast genome. Mitochondrial DNA Part B. 5(2):1868–1869.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Shelke RG, Banerjee RP, Joshi B, Singh PP, Tiwari GJ, Adhikari D, Jena SN, Barik SK. 2022. Chloroplast genome of Lithocarpus dealbatus (Hook. f. & Thomson ex Miq.) Rehder establishes monophyletic origin of the species and reveals mutational hotspots with taxon delimitation potential. Life. 12(6):828.
- Wang MK, Liu X, Zhang ZJ, Yu JW, Liu JJ, Wu YQ. 2021. Phytochemicals and bioactive analysis of different sweet tea (Lithocarpus litseifolius [Hance] Chun) varieties. J Food Biochem. 45(3):e13183.
- Wu CY, Lin L, Yao KP, Yang RJ, Deng M. 2022. The complete chloroplast genome sequence of Lithocarpus longinux (Fagaceae). Mitochondrial DNA B Resour. 7(7):1229–1231.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14(5):1024–1031.
- Zhang SJ, Zhu CC, Bai XQ, Li MM, Chen Y, Zhao YQ. 2021. The complete chloroplast genome of Castanea mollissima ‘Chuizhili. Mitochondrial DNA B Resour. 6(3):1160–1161.
